.jpg.webp)
| Archaeal/bacterial/fungal rhodopsins | |||||||||
|---|---|---|---|---|---|---|---|---|---|
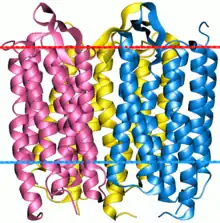 Bacteriorhodopsin trimer | |||||||||
| Identifiers | |||||||||
| Symbol | Bac_rhodopsin | ||||||||
| Pfam | PF01036 | ||||||||
| InterPro | IPR001425 | ||||||||
| SMART | SM01021 | ||||||||
| PROSITE | PDOC00291 | ||||||||
| SCOP2 | 2brd / SCOPe / SUPFAM | ||||||||
| TCDB | 3.E.1 | ||||||||
| OPM superfamily | 6 | ||||||||
| OPM protein | 1vgo | ||||||||
| |||||||||
Microbial rhodopsins, also known as bacterial rhodopsins, are retinal-binding proteins that provide light-dependent ion transport and sensory functions in halophilic[2][3] and other bacteria. They are integral membrane proteins with seven transmembrane helices, the last of which contains the attachment point (a conserved lysine) for retinal.
This protein family includes light-driven proton pumps, ion pumps and ion channels, as well as light sensors. For example, the proteins from halobacteria include bacteriorhodopsin and archaerhodopsin, which are light-driven proton pumps; halorhodopsin, a light-driven chloride pump; and sensory rhodopsin, which mediates both photoattractant (in the red) and photophobic (in the ultra-violet) responses. Proteins from other bacteria include proteorhodopsin.
As their name indicates, microbial rhodopsins are found in Archaea and Bacteria, and also in Eukaryota (such as algae) and viruses; although they are rare in complex multicellular organisms.[4][5]
Nomenclature
Rhodopsin was originally a synonym for "visual purple", a visual pigment (light-sensitive molecule) found in the retinas of frogs and other vertebrates, used for dim-light vision, and usually found in rod cells. This is still the meaning of rhodopsin in the narrow sense, any protein evolutionarily homologous to this protein. In a broad non-genetic sense, rhodopsin refers to any molecule, whether related by genetic descent or not (mostly not), consisting of an opsin and a chromophore (generally a variant of retinal). All animal rhodopsins arose (by gene duplication and divergence) late in the history of the large G-protein coupled receptor (GPCR) gene family, which itself arose after the divergence of plants, fungi, choanoflagellates and sponges from the earliest animals. The retinal chromophore is found solely in the opsin branch of this large gene family, meaning its occurrence elsewhere represents convergent evolution, not homology. Microbial rhodopsins are, by sequence, very different from any of the GPCR families.[6]
The term bacterial rhodopsin originally referred to the first microbial rhodopsin discovered, known today as bacteriorhodopsin. The first bacteriorhodopsin turned out to be of archaeal origin, from Halobacterium salinarum.[7] Since then, other microbial rhodopsins have been discovered, rendering the term bacterial rhodopsin ambiguous.[8][9]
Table
Below is a list of some of the more well-known microbial rhodopsins and some of their properties.
| Function | Name | Abbr. | Ref. |
|---|---|---|---|
| proton pump (H+) | bacteriorhodopsin | BR | [10] |
| proton pump (H+) | proteorhodopsin | PR | [10] |
| proton pump (H+) | archaerhodopsin | Arch | [11] |
| proton pump (H+) | xanthorhodopsin | xR | [12] |
| proton pump (H+) | Gloeobacter rhodopsin | GR | [13] |
| cation channel (+) | channelrhodopsin | ChR | [14] |
| cation pump (Na+) | Krokinobacter eikastus rhodopsin 2 | KR2 | [15] |
| anion pump (Cl-) | halorhodopsin | HR | [10] |
| photosensor | sensory rhodopsin I | SR-I | [10] |
| photosensor | sensory rhodopsin II | SR-II | [10] |
| photosensor | Neurospora opsin I | NOP-I | [14][16] |
| light-activated enzyme | rhodopsin guanylyl cyclase | RhGC | [17] |
The Ion-Translocating Microbial Rhodopsin Family
The Ion-translocating Microbial Rhodopsin (MR) Family (TC# 3.E.1) is a member of the TOG Superfamily of secondary carriers. Members of the MR family catalyze light-driven ion translocation across microbial cytoplasmic membranes or serve as light receptors. Most proteins of the MR family are all of about the same size (250-350 amino acyl residues) and possess seven transmembrane helical spanners with their N-termini on the outside and their C-termini on the inside. There are 9 subfamilies in the MR family:[18]
- Bacteriorhodopsins pump protons out of the cell;
- Halorhodopsins pump chloride (and other anions such as bromide, iodide and nitrate) into the cell;
- Sensory rhodopsins, which normally function as receptors for phototactic behavior, are capable of pumping protons out of the cell if dissociated from their transducer proteins;
- the Fungal Chaperones are stress-induced proteins of ill-defined biochemical function, but this subfamily also includes a H+-pumping rhodopsin;[19]
- the bacterial rhodopsin, called Proteorhodopsin, is a light-driven proton pump that functions as does bacteriorhodopsins;
- the Neurospora crassa retinal-containing receptor serves as a photoreceptor (Neurospora ospin I);[20]
- the green algal light-gated proton channel, Channelrhodopsin-1;
- Sensory rhodopsins from cyanobacteria.
- Light-activated rhodopsin/guanylyl cyclase
A phylogenetic analysis of microbial rhodopsins and a detailed analysis of potential examples of horizontal gene transfer have been published.[21]
Structure
Among the high resolution structures for members of the MR Family are the archaeal proteins, bacteriorhodopsin,[22] archaerhodopsin,[23] sensory rhodopsin II,[24] halorhodopsin,[25] as well as an Anabaena cyanobacterial sensory rhodopsin (TC# 3.E.1.1.6)[26] and others.
Function
The association of sensory rhodopsins with their transducer proteins appears to determine whether they function as transporters or receptors. Association of a sensory rhodopsin receptor with its transducer occurs via the transmembrane helical domains of the two interacting proteins. There are two sensory rhodopsins in any one halophilic archaeon, one (SRI) that responds positively to orange light but negatively to blue light, the other (SRII) that responds only negatively to blue light. Each transducer is specific for its cognate receptor. An x-ray structure of SRII complexed with its transducer (HtrII) at 1.94 Å resolution is available (1H2S).[27] Molecular and evolutionary aspects of the light-signal transduction by microbial sensory receptors have been reviewed.[28]
Homologues
Homologues include putative fungal chaperone proteins, a retinal-containing rhodopsin from Neurospora crassa,[29] a H+-pumping rhodopsin from Leptosphaeria maculans,[19] retinal-containing proton pumps isolated from marine bacteria,[30] a green light-activated photoreceptor in cyanobacteria that does not pump ions and interacts with a small (14 kDa) soluble transducer protein [26][31] and light-gated H+ channels from the green alga, Chlamydomonas reinhardtii.[32] The N. crassa NOP-1 protein exhibits a photocycle and conserved H+ translocation residues that suggest that this putative photoreceptor is a slow H+ pump.[19][33][34]
Most of the MR family homologues in yeast and fungi are of about the same size and topology as the archaeal proteins (283-344 amino acyl residues; 7 putative transmembrane α-helical segments), but they are heat shock- and toxic solvent-induced proteins of unknown biochemical function. They have been suggested to function as pmf-driven chaperones that fold extracellular proteins, but only indirect evidence supports this postulate.[20] The MR family is distantly related to the 7 TMS LCT family (TC# 2.A.43).[20] Representative members of MR family can be found in the Transporter Classification Database.
Bacteriorhodopsin
Bacteriorhodopsin pumps one H+ ion, from the cytosol to the extracellular medium, per photon absorbed. Specific transport mechanisms and pathways have been proposed.[25][35][36] The mechanism involves:
- photo-isomerization of the retinal and its initial configurational changes,
- deprotonation of the retinal Schiff base and the coupled release of a proton to the extracellular membrane surface,
- the switch event that allows reprotonation of the Schiff base from the cytoplasmic side.
Six structural models describe the transformations of the retinal and its interaction with water 402, Asp85, and Asp212 in atomic detail, as well as the displacements of functional residues farther from the Schiff base. The changes provide rationales for how relaxation of the distorted retinal causes movements of water and protein atoms that result in vectorial proton transfers to and from the Schiff base.[35] Helix deformation is coupled to vectorial proton transport in the photocycle of bacteriorhodopsin.[37]
Most residues participating in the trimerization are not conserved in bacteriorhodopsin, a homologous protein capable of forming a trimeric structure in the absence of bacterioruberin. Despite a large alteration in the amino acid sequence, the shape of the intratrimer hydrophobic space filled by lipids is highly conserved between archaerhodopsin-2 and bacteriorhodopsin. Since a transmembrane helix facing this space undergoes a large conformational change during the proton pumping cycle, it is feasible that trimerization is an important strategy to capture special lipid components that are relevant to the protein activity.[38]
Archaerhodopsin
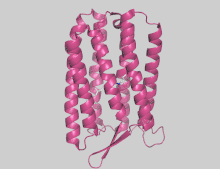
Archaerhodopsins are light-driven H+ ion transporters. They differ from bacteriorhodopsin in that the claret membrane, in which they are expressed, includes bacterioruberin, a second chromophore thought to protect against photobleaching. Bacteriorhodopsin also lacks the omega loop structure that has been observed at the N-terminus of the structures of several archaerhodopsins.
Archaerhodopsin-2 (AR2) is found in the claret membrane of Halorubrum sp. It is a light-driven proton pump. Trigonal and hexagonal crystals revealed that trimers are arranged on a honeycomb lattice.[38] In these crystals, bacterioruberin binds to crevices between the subunits of the trimer. The polyene chain of the second chromophore is inclined from the membrane normal by an angle of about 20 degrees and, on the cytoplasmic side, it is surrounded by helices AB and DE of neighboring subunits. This peculiar binding mode suggests that bacterioruberin plays a structural role for the trimerization of AR2. When compared with the aR2 structure in another crystal form containing no bacterioruberin, the proton release channel takes a more closed conformation in the P321 or P6(3) crystal; i.e., the native conformation of protein is stabilized in the trimeric protein-bacterioruberin complex.
Mutants of Archaerhodopsin-3 (AR3) are widely used as tools in optogenetics for neuroscience research.[39]
Channelrhodopsins
Channelrhodopsin-1 (ChR1) or channelopsin-1 (Chop1; Cop3; CSOA) of C. reinhardtii is closely related to the archaeal sensory rhodopsins. It has 712 aas with a signal peptide, followed by a short amphipathic region, and then a hydrophobic N-terminal domain with seven probable TMSs (residues 76-309) followed by a long hydrophilic C-terminal domain of about 400 residues. Part of the C-terminal hydrophilic domain is homologous to intersection (EH and SH3 domain protein 1A) of animals (AAD30271).
Chop1 serves as a light-gated proton channel and mediates phototaxis and photophobic responses in green algae.[32] Based on this phenotype, Chop1 could be assigned to TC category #1.A, but because it belongs to a family in which well-characterized homologues catalyze active ion transport, it is assigned to the MR family. Expression of the chop1 gene, or a truncated form of that gene encoding only the hydrophobic core (residues 1-346 or 1–517) in frog oocytes in the presence of all-trans retinal produces a light-gated conductance that shows characteristics of a channel passively but selectively permeable to protons. This channel activity probably generates bioelectric currents.[32]
A homologue of ChR1 in C. reinhardtii is channelrhodopsin-2 (ChR2; Chop2; Cop4; CSOB). This protein is 57% identical, 10% similar to ChR1. It forms a cation-selective ion channel activated by light absorption. It transports both monovalent and divalent cations. It desensitizes to a small conductance in continuous light. Recovery from desensitization is accelerated by extracellular H+ and a negative membrane potential. It may be a photoreceptor for dark adapted cells.[40] A transient increase in hydration of transmembrane α-helices with a t(1/2) = 60 μs tallies with the onset of cation permeation. Aspartate 253 accepts the proton released by the Schiff base (t(1/2) = 10 μs), with the latter being reprotonated by aspartic acid 156 (t(1/2) = 2 ms). The internal proton acceptor and donor groups, corresponding to D212 and D115 in bacteriorhodopsin, are clearly different from other microbial rhodopsins, indicating that their spatial positions in the protein were relocated during evolution. E90 deprotonates exclusively in the nonconductive state. The observed proton transfer reactions and the protein conformational changes relate to the gating of the cation channel.[41]
Halorhodopsins
Bacteriorhodopsin pumps one Cl− ion, from the extracellular medium into the cytosol, per photon absorbed. Although the ions move in the opposite direction, the current generated (as defined by the movement of positive charge) is the same as for bacteriorhodopsin and the archaerhodopsins.
Marine Bacterial Rhodopsin
A marine bacterial rhodopsin has been reported to function as a proton pump. However, it also resembles sensory rhodopsin II of archaea as well as an Orf from the fungus Leptosphaeria maculans (AF290180). These proteins exhibit 20-30% identity with each other.
Transport Reaction
The generalized transport reaction for bacterio- and sensory rhodopsins is:[18]
- H+ (in) + hν → H+ (out).
That for halorhodopsin is:
- Cl− (out) + hν → Cl− (in).
See also
References
- ↑ Oren A (January 2002). "Molecular ecology of extremely halophilic Archaea and Bacteria". FEMS Microbiology Ecology. 39 (1): 1–7. Bibcode:2002FEMME..39....1O. doi:10.1111/j.1574-6941.2002.tb00900.x. PMID 19709178.
- ↑ Oesterhelt D, Tittor J (February 1989). "Two pumps, one principle: light-driven ion transport in halobacteria". Trends in Biochemical Sciences. 14 (2): 57–61. doi:10.1016/0968-0004(89)90044-3. PMID 2468194.
- ↑ Blanck A, Oesterhelt D, Ferrando E, Schegk ES, Lottspeich F (December 1989). "Primary structure of sensory rhodopsin I, a prokaryotic photoreceptor". The EMBO Journal. 8 (13): 3963–71. doi:10.1002/j.1460-2075.1989.tb08579.x. PMC 401571. PMID 2591367.
- ↑ Boeuf D, Audic S, Brillet-Guéguen L, Caron C, Jeanthon C (2015). "MicRhoDE: a curated database for the analysis of microbial rhodopsin diversity and evolution". Database. 2015: bav080. doi:10.1093/database/bav080. PMC 4539915. PMID 26286928.
- ↑ Yawo H, Kandori H, Koizumi A (5 June 2015). Optogenetics: Light-Sensing Proteins and Their Applications. Springe r. pp. 3–4. ISBN 978-4-431-55516-2. Retrieved 30 September 2015.
- ↑ Nordström KJ, Sällman Almén M, Edstam MM, Fredriksson R, Schiöth HB (September 2011). "Independent HHsearch, Needleman--Wunsch-based, and motif analyses reveal the overall hierarchy for most of the G protein-coupled receptor families". Molecular Biology and Evolution. 28 (9): 2471–80. doi:10.1093/molbev/msr061. PMID 21402729.
- ↑ Grote M, O'Malley MA (November 2011). "Enlightening the life sciences: the history of halobacterial and microbial rhodopsin research". FEMS Microbiology Reviews. 35 (6): 1082–99. doi:10.1111/j.1574-6976.2011.00281.x. PMID 21623844.
- ↑ "rhodopsin, n.". OED Online. Oxford University Press. 19 December 2012.
- ↑ Mason P (26 May 2011). Medical Neurobiology. OUP USA. p. 375. ISBN 978-0-19-533997-0. Retrieved 21 September 2015.
- 1 2 3 4 5 Yoshizawa S, Kumagai Y, Kim H, Ogura Y, Hayashi T, Iwasaki W, et al. (May 2014). "Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria". Proceedings of the National Academy of Sciences of the United States of America. 111 (18): 6732–7. Bibcode:2014PNAS..111.6732Y. doi:10.1073/pnas.1403051111. PMC 4020065. PMID 24706784.
- ↑ Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, et al. (December 2011). "The microbial opsin family of optogenetic tools". Cell. 147 (7): 1446–57. doi:10.1016/j.cell.2011.12.004. PMC 4166436. PMID 22196724.
- ↑ Sudo Y, Ihara K, Kobayashi S, Suzuki D, Irieda H, Kikukawa T, et al. (February 2011). "A microbial rhodopsin with a unique retinal composition shows both sensory rhodopsin II and bacteriorhodopsin-like properties". The Journal of Biological Chemistry. 286 (8): 5967–76. doi:10.1074/jbc.M110.190058. PMC 3057805. PMID 21135094.
- ↑ Morizumi T, Ou WL, Van Eps N, Inoue K, Kandori H, Brown LS, Ernst OP (August 2019). "X-ray Crystallographic Structure and Oligomerization of Gloeobacter Rhodopsin". Scientific Reports. 9 (1): 11283. Bibcode:2019NatSR...911283M. doi:10.1038/s41598-019-47445-5. PMC 6677831. PMID 31375689. S2CID 199389292.
- 1 2 Heintzen C (2012). "Plant and fungal photopigments". Wiley Interdisciplinary Reviews: Membrane Transport and Signaling. 1 (4): 411–432. doi:10.1002/wmts.36. ISSN 2190-460X.
- ↑ Kato HE, Inoue K, Abe-Yoshizumi R, Kato Y, Ono H, Konno M, et al. (May 2015). "Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump". Nature. 521 (7550): 48–53. Bibcode:2015Natur.521...48K. doi:10.1038/nature14322. PMID 25849775. S2CID 4451644.
- ↑ Olmedo M, Ruger-Herreros C, Luque EM, Corrochano LM (2013). "Regulation of transcription by light in Neurospora crassa: A model for fungal photobiology?". Fungal Biology Reviews. 27 (1): 10–18. doi:10.1016/j.fbr.2013.02.004. ISSN 1749-4613.
- ↑ Scheib U, Stehfest K, Gee CE, Körschen HG, Fudim R, Oertner TG, Hegemann P (August 2015). "The rhodopsin-guanylyl cyclase of the aquatic fungus Blastocladiella emersonii enables fast optical control of cGMP signaling". Science Signaling. 8 (389): rs8. doi:10.1126/scisignal.aab0611. PMID 26268609. S2CID 13140205.
- 1 2 Saier Jr MH. "3.E.1 The Ion-translocating Microbial Rhodopsin (MR) Family". Transporter Classification Database. Saier Lab Bioinformatics Group / SDSC.
- 1 2 3 Waschuk SA, Bezerra AG, Shi L, Brown LS (May 2005). "Leptosphaeria rhodopsin: bacteriorhodopsin-like proton pump from a eukaryote". Proceedings of the National Academy of Sciences of the United States of America. 102 (19): 6879–83. Bibcode:2005PNAS..102.6879W. doi:10.1073/pnas.0409659102. PMC 1100770. PMID 15860584.
- 1 2 3 Zhai Y, Heijne WH, Smith DW, Saier MH (April 2001). "Homologues of archaeal rhodopsins in plants, animals and fungi: structural and functional predications for a putative fungal chaperone protein". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1511 (2): 206–23. doi:10.1016/s0005-2736(00)00389-8. PMID 11286964. S2CID 7931370.
- ↑ Sharma AK, Spudich JL, Doolittle WF (November 2006). "Microbial rhodopsins: functional versatility and genetic mobility". Trends in Microbiology. 14 (11): 463–9. doi:10.1016/j.tim.2006.09.006. PMID 17008099.
- ↑ Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK (October 1999). "Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution". Science. 286 (5438): 255–61. doi:10.1126/science.286.5438.255. PMID 10514362.
- 1 2 Bada Juarez JF, Judge PJ, Adam S, Axford D, Vinals J, Birch J, et al. (January 2021). "Structures of the archaerhodopsin-3 transporter reveal that disordering of internal water networks underpins receptor sensitization". Nature Communications. 12 (1): 629. Bibcode:2021NatCo..12..629B. doi:10.1038/s41467-020-20596-0. PMC 7840839. PMID 33504778.
- ↑ Royant A, Nollert P, Edman K, Neutze R, Landau EM, Pebay-Peyroula E, Navarro J (August 2001). "X-ray structure of sensory rhodopsin II at 2.1-A resolution". Proceedings of the National Academy of Sciences of the United States of America. 98 (18): 10131–6. Bibcode:2001PNAS...9810131R. doi:10.1073/pnas.181203898. PMC 56927. PMID 11504917.
- 1 2 Kolbe M, Besir H, Essen LO, Oesterhelt D (May 2000). "Structure of the light-driven chloride pump halorhodopsin at 1.8 A resolution". Science. 288 (5470): 1390–6. Bibcode:2000Sci...288.1390K. doi:10.1126/science.288.5470.1390. PMID 10827943.
- 1 2 Vogeley L, Sineshchekov OA, Trivedi VD, Sasaki J, Spudich JL, Luecke H (November 2004). "Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 A". Science. 306 (5700): 1390–3. Bibcode:2004Sci...306.1390V. doi:10.1126/science.1103943. PMC 5017883. PMID 15459346.
- ↑ Gordeliy VI, Labahn J, Moukhametzianov R, Efremov R, Granzin J, Schlesinger R, et al. (October 2002). "Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex". Nature. 419 (6906): 484–7. Bibcode:2002Natur.419..484G. doi:10.1038/nature01109. PMID 12368857. S2CID 4425659.
- ↑ Inoue K, Tsukamoto T, Sudo Y (May 2014). "Molecular and evolutionary aspects of microbial sensory rhodopsins". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1837 (5): 562–77. doi:10.1016/j.bbabio.2013.05.005. PMID 23732219.
- ↑ Maturana A, Arnaudeau S, Ryser S, Banfi B, Hossle JP, Schlegel W, et al. (August 2001). "Heme histidine ligands within gp91(phox) modulate proton conduction by the phagocyte NADPH oxidase". The Journal of Biological Chemistry. 276 (32): 30277–84. doi:10.1074/jbc.M010438200. PMID 11389135.
- ↑ Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, et al. (September 2000). "Bacterial rhodopsin: evidence for a new type of phototrophy in the sea". Science. 289 (5486): 1902–6. Bibcode:2000Sci...289.1902B. doi:10.1126/science.289.5486.1902. PMID 10988064.
- ↑ Jung KH, Trivedi VD, Spudich JL (March 2003). "Demonstration of a sensory rhodopsin in eubacteria". Molecular Microbiology. 47 (6): 1513–22. doi:10.1046/j.1365-2958.2003.03395.x. PMID 12622809. S2CID 12052542.
- 1 2 3 Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P (June 2002). "Channelrhodopsin-1: a light-gated proton channel in green algae". Science. 296 (5577): 2395–8. Bibcode:2002Sci...296.2395N. doi:10.1126/science.1072068. PMID 12089443. S2CID 206506942.
- ↑ Brown LS, Dioumaev AK, Lanyi JK, Spudich EN, Spudich JL (August 2001). "Photochemical reaction cycle and proton transfers in Neurospora rhodopsin". The Journal of Biological Chemistry. 276 (35): 32495–505. doi:10.1074/jbc.M102652200. PMID 11435422.
- ↑ Brown LS (June 2004). "Fungal rhodopsins and opsin-related proteins: eukaryotic homologues of bacteriorhodopsin with unknown functions". Photochemical & Photobiological Sciences. 3 (6): 555–65. doi:10.1039/b315527g. PMID 15170485.
- 1 2 Lanyi JK, Schobert B (April 2003). "Mechanism of proton transport in bacteriorhodopsin from crystallographic structures of the K, L, M1, M2, and M2' intermediates of the photocycle". Journal of Molecular Biology. 328 (2): 439–50. doi:10.1016/s0022-2836(03)00263-8. PMID 12691752.
- ↑ Schobert B, Brown LS, Lanyi JK (July 2003). "Crystallographic structures of the M and N intermediates of bacteriorhodopsin: assembly of a hydrogen-bonded chain of water molecules between Asp-96 and the retinal Schiff base". Journal of Molecular Biology. 330 (3): 553–70. doi:10.1016/s0022-2836(03)00576-x. PMID 12842471.
- ↑ Royant A, Edman K, Ursby T, Pebay-Peyroula E, Landau EM, Neutze R (August 2000). "Helix deformation is coupled to vectorial proton transport in the photocycle of bacteriorhodopsin". Nature. 406 (6796): 645–8. Bibcode:2000Natur.406..645R. doi:10.1038/35020599. PMID 10949307. S2CID 4345380.
- 1 2 Yoshimura K, Kouyama T (February 2008). "Structural role of bacterioruberin in the trimeric structure of archaerhodopsin-2". Journal of Molecular Biology. 375 (5): 1267–81. doi:10.1016/j.jmb.2007.11.039. PMID 18082767.
- ↑ Flytzanis NC, Bedbrook CN, Chiu H, Engqvist MK, Xiao C, Chan KY, et al. (September 2014). "Archaerhodopsin variants with enhanced voltage-sensitive fluorescence in mammalian and Caenorhabditis elegans neurons". Nature Communications. 5: 4894. Bibcode:2014NatCo...5.4894F. doi:10.1038/ncomms5894. PMC 4166526. PMID 25222271.
- ↑ Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, et al. (November 2003). "Channelrhodopsin-2, a directly light-gated cation-selective membrane channel". Proceedings of the National Academy of Sciences of the United States of America. 100 (24): 13940–5. Bibcode:2003PNAS..10013940N. doi:10.1073/pnas.1936192100. PMC 283525. PMID 14615590.
- ↑ Lórenz-Fonfría VA, Resler T, Krause N, Nack M, Gossing M, Fischer von Mollard G, et al. (April 2013). "Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating". Proceedings of the National Academy of Sciences of the United States of America. 110 (14): E1273-81. Bibcode:2013PNAS..110E1273L. doi:10.1073/pnas.1219502110. PMC 3619329. PMID 23509282.