| Adenylosuccinate synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
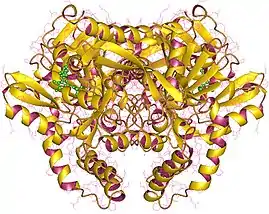 Adenylosuccinate synthetase dimer, Human | |||||||||
| Identifiers | |||||||||
| EC no. | 6.3.4.4 | ||||||||
| CAS no. | 9023-57-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Adenylsucc_synt | |||||||||
|---|---|---|---|---|---|---|---|---|---|
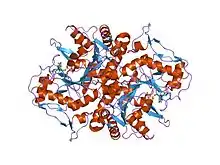 structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana | |||||||||
| Identifiers | |||||||||
| Symbol | Adenylsucc_synt | ||||||||
| Pfam | PF00709 | ||||||||
| Pfam clan | CL0023 | ||||||||
| InterPro | IPR001114 | ||||||||
| PROSITE | PDOC00444 | ||||||||
| SCOP2 | 1ade / SCOPe / SUPFAM | ||||||||
| |||||||||
In molecular biology, adenylosuccinate synthase (or adenylosuccinate synthetase) (EC 6.3.4.4) is an enzyme that plays an important role in purine biosynthesis, by catalysing the guanosine triphosphate (GTP)-dependent conversion of inosine monophosphate (IMP) and aspartic acid to guanosine diphosphate (GDP), phosphate and N(6)-(1,2-dicarboxyethyl)-AMP. Adenylosuccinate synthetase has been characterised from various sources ranging from Escherichia coli (gene purA) to vertebrate tissues. In vertebrates, two isozymes are present: one involved in purine biosynthesis and the other in the purine nucleotide cycle.
Structure
The crystal structure of adenylosuccinate synthetase from E. coli reveals that the dominant structural element of each monomer of the homodimer is a central beta-sheet of 10 strands. The first nine strands of the sheet are mutually parallel with right-handed crossover connections between the strands. The 10th strand is antiparallel with respect to the first nine strands. In addition, the enzyme has two antiparallel beta-sheets, composed of two strands and three strands each, 11 alpha-helices and two short 310-helices. Further, it has been suggested that the similarities in the GTP-binding domains of the synthetase and the p21ras protein are an example of convergent evolution of two distinct families of GTP-binding proteins.[1] Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana when compared with the known structures from E. coli reveals that the overall fold is very similar to that of the E. coli protein.[2]
Isozymes
Humans express two adenylosuccinate synthase isozymes:
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
External links
- Adenylosuccinate+synthase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
References
- ↑ Poland BW, Silva MM, Serra MA, Cho Y, Kim KH, Harris EM, Honzatko RB (December 1993). "Crystal structure of adenylosuccinate synthetase from Escherichia coli. Evidence for convergent evolution of GTP-binding domains". J. Biol. Chem. 268 (34): 25334–42. doi:10.1016/S0021-9258(19)74396-8. PMID 8244965.
- ↑ Prade L, Cowan-Jacob SW, Chemla P, Potter S, Ward E, Fonne-Pfister R (February 2000). "Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana". J. Mol. Biol. 296 (2): 569–77. doi:10.1006/jmbi.1999.3473. PMID 10669609.