Nucleotide sugars are the activated forms of monosaccharides. Nucleotide sugars act as glycosyl donors in glycosylation reactions. Those reactions are catalyzed by a group of enzymes called glycosyltransferases.
History
The anabolism of oligosaccharides - and, hence, the role of nucleotide sugars - was not clear until the 1950s when Leloir and his coworkers found that the key enzymes in this process are the glycosyltransferases. These enzymes transfer a glycosyl group from a sugar nucleotide to an acceptor.[1]
Biological importance and energetics
To act as glycosyl donors, those monosaccharides should exist in a highly energetic form. This occurs as a result of a reaction between nucleoside triphosphate (NTP) and glycosyl monophosphate (phosphate at anomeric carbon). The recent discovery of the reversibility of many glycosyltransferase-catalyzed reactions calls into question the designation of sugar nucleotides as 'activated' donors.[2][3][4][5][6]

Types
There are nine sugar nucleotides in humans which act as glycosyl donors and they can be classified depending on the type of the nucleoside forming them:[7]
- Uridine Diphosphate: UDP-α-D-Glc, UDP-α-D-Gal, UDP-α-D-GalNAc, UDP-α-D-GlcNAc, UDP-α-D-GlcA, UDP-α-D-Xyl
- Guanosine Diphosphate: GDP-α-D-Man, GDP-β-L-Fuc.
- Cytidine Monophosphate: CMP-β-D-Neu5Ac; in humans, it is the only nucleotide sugar in the form of nucleotide monophosphate.
- Cytidine Diphosphate: CDP-D-Ribitol (i.e. CMP-[ribitol phosphate]);[8] though not a sugar, the phosphorylated sugar alcohol ribitol phosphate is incorporated into matriglycan as if it were a monosaccharide.
In other forms of life many other sugars are used and various donors are utilized for them. All five of the common nucleosides are used as a base for a nucleotide sugar donor somewhere in nature. As examples, CDP-glucose and TDP-glucose give rise to various other forms of CDP and TDP-sugar donor nucleotides.[9][10]
Structures
Listed below are the structures of some nucleotide sugars (one example from each type).
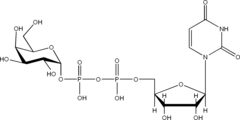 |
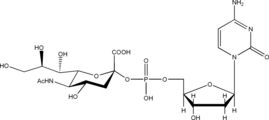 |
 |
| UDP-Gal | CMP-Neu5Ac | GDP-Man |
Relationship to disease
Normal metabolism of nucleotide sugars is very important. Any malfunction in any contributing enzyme will lead to a certain disease [11] for example:
- Inclusion body myopathy: is a congenital disease resulted from altered function of UDP-GlcNAc epimerase .
- Macular corneal dystrophy: is a congenital disease resulted from malfunction of GlcNAc-6-sulfotransferase.
- Congenital disorder in α-1,3 mannosyl transferase will result in a variety of clinical symptoms, e.g. hypotonia, psychomotor retardation, liver fibrosis and various feeding problems.
Relationship to drug discovery
The development of chemoenzymatic strategies to generate large libraries of non-native sugar nucleotides has enabled a process referred to as glycorandomization where these sugar nucleotide libraries serve as donors for permissive glycosyltransferases to afford differential glycosylation of a wide range of pharmaceuticals and complex natural product-based leads.[12][13]
See also
References
- ↑ Derek Horton (2008). "The Development of Carbohydrate Chemistry and Biology". Carbohydrate Chemistry, Biology and Medical Applications: 1–28. doi:10.1016/B978-0-08-054816-6.00001-X. ISBN 978-0-08-054816-6.
- ↑ Zhang, C; Griffith, BR; Fu, Q; Albermann, C; Fu, X; Lee, IK; Li, L; Thorson, JS (1 September 2006). "Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions". Science. 313 (5791): 1291–4. doi:10.1126/science.1130028. PMID 16946071. S2CID 38072017.
- ↑ Zhang, C; Albermann, C; Fu, X; Thorson, JS (27 December 2006). "The in vitro characterization of the iterative avermectin glycosyltransferase AveBI reveals reaction reversibility and sugar nucleotide flexibility". Journal of the American Chemical Society. 128 (51): 16420–1. doi:10.1021/ja065950k. PMID 17177349.
- ↑ Zhang, C; Fu, Q; Albermann, C; Li, L; Thorson, JS (5 March 2007). "The in vitro characterization of the erythronolide mycarosyltransferase EryBV and its utility in macrolide diversification". ChemBioChem. 8 (4): 385–90. doi:10.1002/cbic.200600509. PMID 17262863. S2CID 45058028.
- ↑ Zhang, C; Moretti, R; Jiang, J; Thorson, JS (13 October 2008). "The in vitro characterization of polyene glycosyltransferases AmphDI and NysDI". ChemBioChem. 9 (15): 2506–14. doi:10.1002/cbic.200800349. PMC 2947747. PMID 18798210.
- ↑ Gantt, RW; Peltier-Pain, P; Cournoyer, WJ; Thorson, JS (21 August 2011). "Using simple donors to drive the equilibria of glycosyltransferase-catalyzed reactions". Nature Chemical Biology. 7 (10): 685–91. doi:10.1038/nchembio.638. PMC 3177962. PMID 21857660.
- ↑ Cold Spring Harbor Laboratory Press Archived 2011-07-08 at the Wayback Machine Essentials of Glycobiology, Second Edition
- ↑ Gerin I, et al. (2016). "ISPD produces CDP-ribitol used by FKTN and FKRP to transfer ribitol phosphate onto α-dystroglycan". Nature Communications. 7: 11534. doi:10.1038/ncomms11534. PMC 4873967. PMID 27194101.
- ↑ Samuel G, Reeves P (2003). "Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly". Carbohydr. Res. 338 (23): 2503–19. doi:10.1016/j.carres.2003.07.009. PMID 14670712.
- ↑ Xue M. He; Hung-wen Liu (2002). "Formation of unusual sugars: Mechanistic studies and biosynthetic applications". Annu Rev Biochem. 71: 701–754. doi:10.1146/annurev.biochem.71.110601.135339. PMID 12045109.
- ↑ Encyclopedia of Biological Chemistry, Volume 2. 2004, Elsevier Inc. Hudson H. Freeze 302-307.
- ↑ Langenhan, JM; Griffith, BR; Thorson, JS (November 2005). "Neoglycorandomization and chemoenzymatic glycorandomization: two complementary tools for natural product diversification". Journal of Natural Products. 68 (11): 1696–711. doi:10.1021/np0502084. PMID 16309329.
- ↑ Gantt, RW; Peltier-Pain, P; Thorson, JS (October 2011). "Enzymatic methods for glyco(diversification/randomization) of drugs and small molecules". Natural Product Reports. 28 (11): 1811–53. doi:10.1039/c1np00045d. PMID 21901218.
External links
- "Nucleotide sugars" at the U.S. National Library of Medicine Medical Subject Headings (MeSH)