| 4-hydroxybenzoate 3-monooxygenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystal structure of 4-hydroxybenzoate 3-monooxygenase | |||||||||
| Identifiers | |||||||||
| EC no. | 1.14.13.2 | ||||||||
| CAS no. | 9059-23-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
The enzyme 4-hydroxybenzoate 3-monooxygenase, also commonly referred to as para-hydroxybenzoate hydroxylase (PHBH), is a flavoprotein belonging to the family of oxidoreductases. Specifically, it is a hydroxylase, and is one of the most studied enzymes and catalyzes reactions involved in soil detoxification, metabolism, and other biosynthetic processes.[1]
4-hydroxybenzoate 3-monooygenase catalyzes the regioselective hydroxylation of 4-hydroxybenzoate, giving 3,4-dihydroxybenzoate as the product. The mechanism consists of the following general steps: (1) reduction of the flavin, (2) reaction of the flavin with O2, producing C4a-hydroperoxyflavin, and (3) binding and activation of the substrate, leading to product formation and release.[2] Throughout the mechanism, the flavin changes between “open” and “closed” conformations, thus altering the substrate reaction environment. The open conformation allows solvent access to the active site; the enzyme adopts this conformation for substrate binding and product release. A closed conformation isolates the reaction from solvent, which helps to stabilize the reaction intermediates.[2]

Structure
4-hydroxybenzoate 3-monooxygenase is a homodimer with a flavin bound to each monomer. The active site is composed of the flavin and amino acids on the monomer. The structure of this enzyme often serves as a model for structure-reactivity interdependence of other flavin-dependent hydroxylases. The active site limits potential substrates to substituted benzenes, namely 4-hydroxybenzoate (the native substrate), 2,4-dihydroxybenzoate, 4-mercaptobenzoate, and several halogenated aromatic compounds.[3]
Mechanism
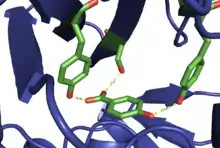
The hydroxylase, 4-hydroxybenzoate 3-monooxygenase, proceeds through a catalytic process that begins with the entrance of NADPH and 4-hydroxybenzoate (the native substrate) into the active site of the enzyme. This results in formation of an enzyme-flavin-substrate-NADPH complex, after which the flavin cofactor, FAD, is reduced by NADPH. NADP+ is lost and O2 enters into the complex, followed by oxidation of the flavin to form a hydroperoxide, which acts as the hydroxide transfer reagent. It is important to note that while the group transferred is referred to as hydroxide it is formally an OH+ group. This hydroxide is transferred to the substrate from the hydroperoxide flavin, flavin-C4a-hydroperoxide, via an electrophilic aromatic substitution-type reaction. Finally, the product exits from the complex and the hydroxy-flavin is dehydrated, regenerating FAD and allowing the process to repeat.[2]
The substrate binds in the active site of the enzyme via non-covalent interactions with proximal amino acid side chains. Specifically, the hydroxyl groups of tyrosine 201 and 222, in addition to the hydroxyl group of serine 212, interact with the carboxylate and hydroxyl group on the substrate. This allows for proper orientation within the active site in order to achieve reactivity.[2]
Once the substrate is bound, the flavin shifts from an “open” to a “closed” conformation. This shields the active site and substrate from solvent, preventing the premature breakdown of the flavin hydroperoxide. Binding of both NADPH and substrate shifts the enzyme to an “out” conformation. This occurs through an intricate proton network within the enzyme that allows for deprotonation of the phenol and a subsequent dynamic shift of the enzyme. The “out” conformation aligns the isoalloxazine ring of the flavin so that it can be reduced rapidly by NADPH. Following the reduction, NADP+ is released from the enzyme.[4]
Reduction of the flavin generates a negatively charged species, FADH−, which is attracted to the positively charged active site. This attraction shifts the flavin back to the “closed” conformation, isolating it from the solvent environment. This isolation provides an optimal environment and position for O2 to hydroxylate the substrate.[4] The oxygen binds to FADH− via a single electron transfer, which is the rate-limiting step of the reaction. This forms an FAD radical and flavin hydroperoxide. Reaction between these generates C4a-peroxyflavin, which is quickly protonated to form flavin-C4a-hydroperoxide.[3] Tautomerization leads to the formation of 3,4-dihydoxybenzoate. The final step in the mechanism is dissociation of the product and water from FAD, causing the flavin to return to the open conformation.[1]
References
- 1 2 Entsch B, Ballou DP, Begley TP (2007-01-01). Wiley Encyclopedia of Chemical Biology. John Wiley & Sons, Inc. doi:10.1002/9780470048672.wecb672. ISBN 9780470048672.
- 1 2 3 4 Gatti DL, Palfey BA, Lah MS, Entsch B, Massey V, Ballou DP, Ludwig ML (October 1994). "The mobile flavin of 4-OH benzoate hydroxylase". Science. 266 (5182): 110–4. Bibcode:1994Sci...266..110G. doi:10.1126/science.7939628. PMID 7939628.
- 1 2 Montersino S, Tischler D, Gassner GT, van Berkel WJ (September 2011). "Catalytic and Structural Features of Flavoprotein Hydroxylases and Epoxidases". Advanced Synthesis & Catalysis. 353 (13): 2301–2319. doi:10.1002/adsc.201100384.
- 1 2 Ballou DP, Entsch B, Cole LJ (December 2005). "Dynamics involved in catalysis by single-component and two-component flavin-dependent aromatic hydroxylases". Biochemical and Biophysical Research Communications. 338 (1): 590–8. doi:10.1016/j.bbrc.2005.09.081. PMID 16236251.
Further reading
- Hosokawa K, Stanier RY (May 1966). "Crystallization and properties of p-hydroxybenzoate hydroxylase from Pseudomonas putida". The Journal of Biological Chemistry. 241 (10): 2453–60. PMID 4380381.
- Howell LG, Spector T, Massey V (July 1972). "Purification and properties of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens". The Journal of Biological Chemistry. 247 (13): 4340–50. PMID 4402514.
- Spector T, Massey V (July 1972). "Studies on the effector specificity of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens". The Journal of Biological Chemistry. 247 (14): 4679–87. PMID 4402938.
- Spector T, Massey V (September 1972). "p-Hydroxybenzoate hydroxylase from Pseudomonas fluorescens. Evidence for an oxygenated flavin intermediate". The Journal of Biological Chemistry. 247 (17): 5632–6. PMID 4403446.
- Spector T, Massey V (November 1972). "p-Hydroxybenzoate hydroxylase from Pseudomonas fluorescens. Reactivity with oxygen". The Journal of Biological Chemistry. 247 (22): 7123–7. PMID 4404745.
- Seibold B, Matthes M, Eppink MH, Lingens F, Van Berkel WJ, Müller R (July 1996). "4-Hydroxybenzoate hydroxylase from Pseudomonas sp. CBS3. Purification, characterization, gene cloning, sequence analysis and assignment of structural features determining the coenzyme specificity". European Journal of Biochemistry. 239 (2): 469–78. doi:10.1111/j.1432-1033.1996.0469u.x. PMID 8706756.