| aspartate racemase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 5.1.1.13 | ||||||||
| CAS no. | 37237-56-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
In enzymology, an aspartate racemase (EC 5.1.1.13) is an enzyme that catalyzes the following chemical reaction:
- L-aspartate D-aspartate
This enzyme belongs to the family of isomerases, specifically those racemases and epimerases acting on amino acids and amino acid derivatives, including glutamate racemase, proline racemase, and diaminopimelate epimerase.
The systematic name of this enzyme class is aspartate racemase. Other names in common use include D-aspartate racemase, and McyF.[1]
Discovery
Aspartate racemase was first discovered in the gram-positive bacteria Streptococcus faecalis by Lamont et al. in 1972.[2] It was then determined that aspartate racemase also racemizes L-alanine around half as quickly as it does L-aspartate, but does not show racemase activity in the presence of L-glutamate.
Structure
The crystallographic structure of bacterial aspartate racemase has been solved in Pyrococcus horikoshii OT3,[3] Escherichia coli, Microcystis aeruginosa, and Picrophilus torridus DSM 9790.
Homodimer
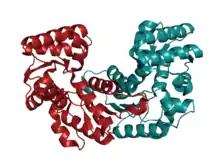
In most bacteria for which the structure is known, aspartate racemase exists as a homodimer,[3] where each subunit has a molecular weight of approximately 25 kDa. The complex consists primarily of alpha helices, and additionally features a Rossmann fold in the center of the dimer. The catalytic pocket lies at the cleft formed by the intersection of the two domains. A citrate molecule can fit inside the binding pocket, leading to a contraction of the cleft to make the "closed form" of aspartate racemase.[4]
Two highly conserved cysteine residues are suggested to be responsible for the interconversion of L-aspartate and D-aspartate.[3] These cysteine residues lie 3–4 angstroms away from the α-carbon of aspartate. Site-directed mutagenesis studies showed that the mutation of the upstream cysteine residue to serine resulted in complete loss of racemization activity, while the same mutation in the downstream cysteine residue resulted in retention of 10–20% racemization activity.[4] However, mutation of the acid residue glutamate, which stabilizes the downstream cysteine residue, resulted in complete loss of racemization activity. Up to 9 other residues are known to interact with and stabilize the isomers of aspartate through hydrogen bonding or hydrophobic interactions.[5][4]
In E. coli, one of the active cysteine residues is substituted for a threonine residue, allowing for much greater substrate promiscuity.[5] Notably, aspartate racemase in E. coli is also able to catalyze the racemization of glutamate.
Monomer
In 2004, an aspartate racemase was discovered in Bifidobacterium bifidum as a 27 kDa monomer.[6] This protein shares nearly identical enzymological properties with homodimeric aspartate racemase isolated from Streptococcus thermophilus, but has the added characteristic that its thermal stability increases significantly in the presence of aspartate.
Reaction mechanism
Aspartate racemase catalyzes the following reaction:
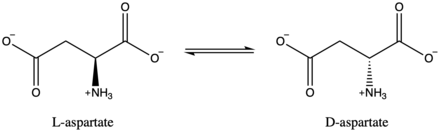
Aspartate racemase can accept either L-aspartate or D-aspartate as substrates.
Amino acid racemization is carried out by two dominant mechanisms: one-base mechanisms and two-base mechanisms.[7] In one-base mechanisms, a proton acceptor abstracts the α-hydrogen from the substrate amino acid to form a carbanion intermediate until reprotonated at the other face of the α-carbon. Racemases dependent on pyridoxal-5-phosphate (PLP) typically leverage one-base mechanisms. In the two-base mechanism, an alpha hydrogen is abstracted by a base on one face of the amino acid while another protonated base concertedly donates its hydrogen onto the other face of the amino acid.
PLP-independent mechanism
Aspartate racemases in bacteria function in the absence of PLP, suggesting a PLP-independent mechanism.[5] A two-base mechanism is supported in the literature, carried out by two thiol groups:

Other PLP-independent isomerases in bacteria include glutamate racemase, proline racemase, and hydroxyproline-2-epimerase.
PLP-dependent mechanism
Mammalian aspartate racemase, in contrast with bacterial aspartate racemase, is a PLP-dependent enzyme. The exact mechanism is unknown, but it is hypothesized to proceed similarly to mammalian serine racemase as below:

Inhibition
General inhibitors for cysteine residues have shown to be effective agents against monomeric aspartate racemase.[2][5] N-ethylmaleimide and 5,5'-dithiobis(2-nitrobenzoate) both inhibit monomeric aspartate racemase at 1mM.
Function
Metabolism of D-aspartate
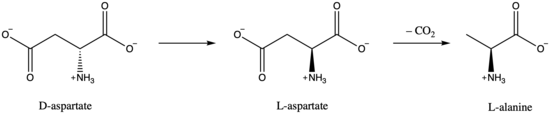
One of the primary functions of aspartate racemase in bacteria is the metabolism of D-aspartate.[8][9] The beginning of D-aspartate metabolism is its conversion to L-alanine. First, D-aspartate is isomerized to L-aspartate by aspartate racemase, followed by decarboxylation to form L-alanine.[9]
Peptidoglycan synthesis
D-amino acids are common within the peptidoglycan of bacteria.[10] In Bifidobacterium bifidum, D-aspartate is formed from L-aspartate via aspartate racemase and used as a cross-linker moiety in the peptidoglycan.[6]
Mammalian neurogenesis
Aspartate racemase is highly expressed in the brain, the heart, and the testes of mammals, all tissues in which D-aspartate is present.[11] D-aspartate is abundant in the embryonic brain, but falls during postnatal development. Retrovirus-mediated expression of short hairpin RNA complementary to aspartate racemase in newborn neurons of the adult hippocampus led to defects in dendritic development and empaired survival of the newborn neurons, suggesting that aspartate racemase may modulate adult neurogenesis in mammals.[12]
Evolution
Phylogenetic analysis shows that PLP-dependent animal aspartate racemases are in the same family as PLP-dependent animal serine racemases, and the genes encoding them share a common ancestor.[13] Aspartate racemases in animals have independently evolved from serine racemases through amino acid substitutions, namely, the introduction of three consecutive serine residues.[14] Serine racemases isolated from Saccoglossus kowalevskii also show both high aspartate and glutamate racemization activity.[15]
References
- ↑ Sielaff H, Dittmann E, Tandeau De Marsac N, Bouchier C, Von Döhren H, Börner T, Schwecke T (August 2003). "The mcyF gene of the microcystin biosynthetic gene cluster from Microcystis aeruginosa encodes an aspartate racemase". The Biochemical Journal. 373 (Pt 3): 909–16. doi:10.1042/BJ20030396. PMC 1223527. PMID 12713441.
- 1 2 Lamont HC, Staudenbauer WL, Strominger JL (August 1972). "Partial purification and characterization of an aspartate racemase from Streptococcus faecalis". The Journal of Biological Chemistry. 247 (16): 5103–6. doi:10.1016/S0021-9258(19)44944-2. PMID 4626916.
- 1 2 3 Liu L, Iwata K, Kita A, Kawarabayasi Y, Yohda M, Miki K (May 2002). "Crystal structure of aspartate racemase from Pyrococcus horikoshii OT3 and its implications for molecular mechanism of PLP-independent racemization". Journal of Molecular Biology. 319 (2): 479–89. doi:10.1016/S0022-2836(02)00296-6. PMID 12051922.
- 1 2 3 Cao DD, Zhang CP, Zhou K, Jiang YL, Tan XF, Xie J, et al. (July 2019). "Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF". Biochemical and Biophysical Research Communications. 514 (4): 1108–1114. doi:10.1016/j.bbrc.2019.05.063. PMID 31101340.
- 1 2 3 4 Fischer C, Ahn YC, Vederas JC (December 2019). "Catalytic mechanism and properties of pyridoxal 5'-phosphate independent racemases: how enzymes alter mismatched acidity and basicity". Natural Product Reports. 36 (12): 1687–1705. doi:10.1039/c9np00017h. PMID 30994146.
- 1 2 Yamashita T, Ashiuchi M, Ohnishi K, Kato S, Nagata S, Misono H (December 2004). "Molecular identification of monomeric aspartate racemase from Bifidobacterium bifidum". European Journal of Biochemistry. 271 (23–24): 4798–803. doi:10.1111/j.1432-1033.2004.04445.x. PMID 15606767.
- ↑ Yamauchi T, Choi SY, Okada H, Yohda M, Kumagai H, Esaki N, Soda K (September 1992). "Properties of aspartate racemase, a pyridoxal 5'-phosphate-independent amino acid racemase". The Journal of Biological Chemistry. 267 (26): 18361–4. doi:10.1016/S0021-9258(19)36969-8. PMID 1526977.
- ↑ Henríquez T, Salazar JC, Marvasi M, Shah A, Corsini G, Toro CS (2020). "SRL pathogenicity island contributes to the metabolism of D-aspartate via an aspartate racemase in Shigella flexneri YSH6000". PLOS ONE. 15 (1): e0228178. Bibcode:2020PLoSO..1528178H. doi:10.1371/journal.pone.0228178. PMC 6980539. PMID 31978153.
- 1 2 Markovetz AJ, Cook WJ, Larson AD (August 1966). "Bacterial metabolism of d-aspartate involving racemization and decarboxylation". Canadian Journal of Microbiology. 12 (4): 745–51. doi:10.1139/m66-101. PMID 5969337.
- ↑ Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK (September 2009). "D-amino acids govern stationary phase cell wall remodeling in bacteria". Science. 325 (5947): 1552–5. Bibcode:2009Sci...325.1552L. doi:10.1126/science.1178123. PMC 2759711. PMID 19762646.
- ↑ Ohide H, Miyoshi Y, Maruyama R, Hamase K, Konno R (November 2011). "D-Amino acid metabolism in mammals: biosynthesis, degradation and analytical aspects of the metabolic study". Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences. 879 (29): 3162–8. doi:10.1016/j.jchromb.2011.06.028. PMID 21757409.
- ↑ Kim PM, Duan X, Huang AS, Liu CY, Ming GL, Song H, Snyder SH (February 2010). "Aspartate racemase, generating neuronal D-aspartate, regulates adult neurogenesis". Proceedings of the National Academy of Sciences of the United States of America. 107 (7): 3175–9. Bibcode:2010PNAS..107.3175K. doi:10.1073/pnas.0914706107. PMC 2840285. PMID 20133766.
- ↑ Uda K, Abe K, Dehara Y, Mizobata K, Sogawa N, Akagi Y, et al. (February 2016). "Distribution and evolution of the serine/aspartate racemase family in invertebrates". Amino Acids. 48 (2): 387–402. doi:10.1007/s00726-015-2092-0. PMID 26352274. S2CID 17216091.
- ↑ Uda K, Abe K, Dehara Y, Mizobata K, Edashige Y, Nishimura R, et al. (October 2017). "Triple serine loop region regulates the aspartate racemase activity of the serine/aspartate racemase family". Amino Acids. 49 (10): 1743–1754. doi:10.1007/s00726-017-2472-8. PMID 28744579. S2CID 3707632.
- ↑ Uda K, Ishizuka N, Edashige Y, Kikuchi A, Radkov AD, Moe LA (June 2019). "Cloning and characterization of a novel aspartate/glutamate racemase from the acorn worm Saccoglossus kowalevskii". Comparative Biochemistry and Physiology. Part B, Biochemistry & Molecular Biology. 232: 87–92. doi:10.1016/j.cbpb.2019.03.006. PMID 30902582.