| Bidensovirus | |
|---|---|
.jpg.webp) | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Monodnaviria |
| Kingdom: | Shotokuvirae |
| Phylum: | Cossaviricota |
| Class: | Mouviricetes |
| Order: | Polivirales |
| Family: | Bidnaviridae |
| Genus: | Bidensovirus |
| Species | |
|
Bombyx mori bidensovirus | |
Bidensovirus is a genus of single stranded DNA viruses that infect invertebrates. The species in this genus were originally classified in the family Parvoviridae (subfamily Densovirinae) but were moved to a new genus because of significant differences in the genomes.[1]
Taxonomy
There is one species in this genus currently recognised: Bombyx mori bidensovirus.
Host
As the name suggests this virus infects Bombyx mori, the silkworm.[2]
Virology
The virions are icosahedral, non enveloped and ~25 nanometers in diameter. They contain two structural proteins.
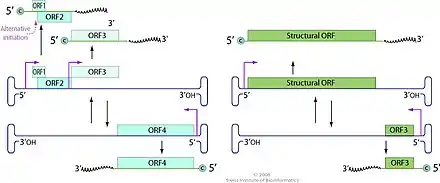
The genome is bipartite, unique among ssDNA viruses, with two linear segments of ~6 and 6.5 kilobases (kb). These segments and the complementary strands are that are packaged separately giving rise to 4 different types of full particles.
Both segments have an ambisense organization, coding for a structural protein in one sense and the non-structural proteins on the complementary strand.
- DNA1 (also known as VD1) — the larger segment of 6.5 kb — encodes the capsid protein VP1 (128 kDa — kilodaltons) on one strand and three non-structural proteins — NS1 of 14 kDa, NS2 of 37 kDa and NS3 of 55 kDa — on the complementary strand.
- DNA2 (also known as VD2) — the smaller segment of 6 kb — encodes the capsid protein VP2 (133 kDa) on one strand and the non-structural protein NS4 (27 kDa) on the complementary strand.
The open reading frame 4 (VD1-ORF4) is 3318 nucleotides (bases) in length and encodes a predicted (3318/3 − 1 =) 1105 amino acid protein which has a conserved DNA polymerase motif. It appears to encode at least 2 other proteins including one of ~53 kDa that forms part of the virion.[3]
Evolution
Comprehensive analysis of bidnavirus genes has shown that these viruses have evolved from a parvovirus ancestor from which they inherit a jelly-roll capsid protein and a superfamily 3 helicase.[4] It has been further suggested that the key event that led to the separation of the bidnaviruses from parvoviruses was the acquisition of the PolB gene. A likely scenario has been proposed under which the ancestral parvovirus genome was integrated into a large virus-derived DNA transposon of the Polinton/Maverick family (polintoviruses) [5] resulting in the acquisition of the polintovirus PolB gene along with terminal inverted repeats. Bidnavirus genes for a minor structural protein (putative receptor-binding protein) and a potential novel antiviral defense modulator were derived from dsRNA viruses (Reoviridae) and dsDNA viruses (Baculoviridae), respectively.[4]
References
- ↑ Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses (2011) ISBN 978-0123846846
- ↑ "Bidensovirus ~ ViralZone page".
- ↑ Li, Guohui; Hu, Zhaoyang; Guo, Xuli; Li, Guangtian; Tang, Qi; Wang, Peng; Chen, Keping; Yao, Qin (June 2013). "Li G, Hu Z, Guo X, Li G, Tang Q, Wang P, Chen K, Yao Q Identification of Bombyx mori Bidensovirus VD1-ORF4 Reveals a Novel Protein Associated with Viral Structural Component. Curr Microbiol 66, 527–534 (2013)". Current Microbiology. 66 (6): 527–534. doi:10.1007/s00284-013-0306-9. PMID 23328902. S2CID 15920465.
- 1 2 Krupovic M, Koonin EV (2014). "Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses". Sci Rep. 4: 5347. doi:10.1038/srep05347. PMC 4061559. PMID 24939392.
- ↑ Krupovic M, Bamford DH, Koonin EV (2014). "Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses". Biol Direct. 9 (1): 6. doi:10.1186/1745-6150-9-6. PMC 4028283. PMID 24773695.
External links
- Bidensovirus ~ ViralZone page. The top image erraneously shows a monopartite genome, text and genome map are right (March 30, 2021).