
Biotin carboxyl carrier protein (BCCP) refers to proteins containing a biotin attachment domain that carry biotin and carboxybiotin throughout the ATP-dependent carboxylation by biotin-dependent carboxylases. The biotin carboxyl carrier protein is an Acetyl CoA subunit that allows for Acetyl CoA to be catalyzed and converted to malonyl-CoA. More specifically, BCCP catalyzes the carboxylation of the carrier protein to form an intermediate. Then the carboxyl group is transferred by the transcacrboxylase to form the malonyl-CoA.[1] This conversion is an essential step in the biosynthesis of fatty acids. In the case of E. coli Acetyl-CoA carboxylase, the BCCP is a separate protein known as accB (P0ABD8). On the other hand, in Haloferax mediterranei, propionyl-CoA carboxylase, the BCCP pccA (I3R7G3) is fused with biotin carboxylase.
The biosynthesis of fatty acids in plants, such as triacylglycerol, is vital to the plant's overall health because it allows for accumulation of seed oil. The biosynthesis that is catalyzed by BCCP usually takes place in the chloroplast of plant cells. The biosynthesis performed by the BCCP protein allows for the transfer of CO2 within active sites of the cell.[2]
The biotin carboxyl carrier protein carries approximately 1 mol of biotin per 22,000 g of protein.[3]
There is not much research on BCCPs at the moment. However, a recent studyon plant genomics found that Brassica BCCPs might play a key role in abiotic and biotic stress responses.[4] Meaning that these proteins may be relaying messages to the rest of the plant body after it has been exposed to extreme conditions that disrupt the plant's homeostasis.
Synthesis of Malonyl-CoA
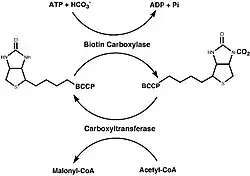 |
The synthesis of Malonyl-CoA consists of two half reactions. The first being the carboxylation of biotin with bicarbonate and the second being the transfer of the CO2 group to acetyl-CoA from carboxybiotin to allow for the formation of malonyl-CoA. Two different protein subassemblies, along with BCCP, are required for this two step reaction to be successful: biotin carboxylase (BC) and carboxyltransferase (CT). BCCP contains the biotin cofactor which is covalently bound to a lysine residue.[5]
In fungi, mammals, and plant cytosols, all three of these components (BCCP, BC, and CT) exist on one polypeptide chain. However, most studies of this protein have been conducted on the E. coli form of the enzyme, where all three components exist as three separate complexes rather than being united on one polypeptide chain.
Structure
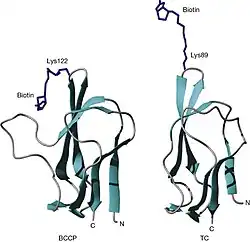 Three-dimensional structures of two biotin domains. The structures of the biotin domains from the biotin carboxyl carrier protein (BCCP) of (left) Escherichia coli acetyl CoA carboxylase and (right) the 1.3S subunit of Propionibacterium freudenreichii subsp. shermanii transcarboxylase (TC). |
The first report of the BCCP structure was made by biochemists F. K. Athappilly and W. A. Hendrickson in 1995.[6] It can be thought of as a long β-hairpin structure, with four pairs of antiparallel β-strands that wrap around a central hydrophobic core. The biotinylation motif Met-Lys-Met is located at the tip of the β-hairpin structure. Rotations around the CαCβ bond of this Lys residue contribute to the swinging-arm model. The connection to the rest of the enzyme at the N-terminus of BCCP core is located at the opposite end of the structure from the biotin moiety. Rotations around this region contribute to the swinging-domain model, and the N1′ atom of biotin is ~ 40 Å from this pivot point. This gives a range of ~ 80 Å for the swinging-domain model, and the BC–CT active site distances observed so far are between 40 and 80 Å.[7] In addition, the linker before the BCCP core in the holoenzyme could also be flexible, which would give further reach for the biotin N1′ atom.[8]
The structures of biotin-accepting domains from E. coli BCCP-87 and the 1.3S subunit of P. shermanii TC were determined by both X-ray crystallography and nuclear magnetic resonance studies. (Athappilly and Hendrickson, 1995; Roberts et al., 1999; Reddy et al., 1998).[9] These produced essentially the same structures that are structurally related to the lipoyl domains of 2-oxo acid dehydrogenase multienzyme complexes (Brocklehurst and Perham, 1993; Dardel et al., 1993), which similarly undergo an analogous post-translational modification. These domains form a flattened β-barrel structure comprising two four-stranded β-sheets with the N- and C-terminal residues close together at one end of the structure. At the other end of the molecule, the biotinyl- or lipoyl-accepting lysine resides on a highly exposed, tight hairpin loop between β4 and β5 strands. The structure of the domain is stabilized by a core of hydrophobic residues, which are important structural determinants. Conserved glycine residues occupy β-turns linking the β-strands.[10]
The structure of the Biotin-accepting domain consists of BCCP-87 which contains a seven-amino-acid insertion common to certain prokaryotic acetyl-CoA carboxylases but not present in other biotindomains (Chapman-Smith and Cronan, 1999). This region of the peptide adopts a thumb structure between the β2 and β3 strands and, interestingly, forms direct contacts with the biotin moiety in both the crystal and solution structures (Athappilly and Hendrickson, 1995; Roberts et al., 1999). It has been proposed that this thumb may function as a mobile lid for either, or possibly both, the biotin carboxylase or carboxyl- transferase active sites in the biotin-dependent enzyme (Cronan, 2001). The function of this lid could aid to prevent solvation of the active sites, thereby aiding in the transfer of CO2 from carboxybiotin to acetyl CoA. Secondly, the thumb is required for dimerization of BCCP, necessary for the formation of the active acetyl CoA carboxylase complex (Cronan, 2001). In conclusion, the thumb functions to inhibit the aberrant lipoylation of the target lysine by lipoyl protein ligase (Reche and Perham, 1999). Removal of the thumb by mutagenesis rendered BCCP-87 a favorable substrate for lipoylation but abolished biotinylation (Reche and Perham, 1999). The thumb structure, however, is not a highly conserved feature amongst all biotin domains. Many biotin-dependent enzymes do not contain this insertion, including all five mammalian enzymes. However, it appears the interactions between biotin and protein might be a conserved feature and important for catalysis as similar contacts have been observed in the "thumbless" domains from P. shermanii transcarboxylase (Jank et al., 2002) and the biotinyl/lipoyl attachment protein of B. subtilis (Cui et al., 2006). The significance of this requires further investigation but it is possible that the mechanism employed by the biotin enzymes may involve noncovalent interactions between the protein and the prosthetic group.
References
- ↑ "UniProt". www.uniprot.org. Retrieved 2023-04-26.
- ↑ "InterPro". www.ebi.ac.uk. Retrieved 2023-04-26.
- ↑ Fall, R. Ray; Nervi, A. M.; Alberts, Alfred W.; Vagelos, P. Roy (July 1971). "Acetyl CoA Carboxylase: Isolation and Characterization of Native Biotin Carboxyl Carrier Protein". Proceedings of the National Academy of Sciences. 68 (7): 1512–1515. Bibcode:1971PNAS...68.1512F. doi:10.1073/pnas.68.7.1512. ISSN 0027-8424. PMC 389229. PMID 4934522.
- ↑ Megha, Swati; Wang, Zhengping; Kav, Nat N. V.; Rahman, Habibur (2022-10-17). "Genome-wide identification of biotin carboxyl carrier subunits of acetyl-CoA carboxylase in Brassica and their role in stress tolerance in oilseed Brassica napus". BMC Genomics. 23 (1): 707. doi:10.1186/s12864-022-08920-y. ISSN 1471-2164. PMC 9578262. PMID 36253756.
- ↑ Choi-Rhee, Eunjoo (August 2003). "The Biotin Carboxylase-Biotin Carboxyl Carrier Protein Complex of Escherichia coli Acetyl-CoA Carboxylase". Journal of Biological Chemistry. 278 (33): 30806–30812. doi:10.1074/jbc.M302507200. PMID 12794081.
- ↑ Athappilly, F. K.; Hendrickson, W. A. (1995-12-15). "Structure of the biotinyl domain of acetyl-coenzyme A carboxylase determined by MAD phasing". Structure. 3 (12): 1407–1419. doi:10.1016/s0969-2126(01)00277-5. ISSN 0969-2126. PMID 8747466. S2CID 21461025.
- ↑ Rossen Donev, ed. (11 April 2023). Control of Cell Cycle & Cell Proliferation. Advances in Protein Chemistry and Structural Biology Vol. 135. Oxford [u.a.]: Academic Press. ISBN 978-0-443-15822-3.
- ↑ Tong, Liang (2017), Striking Diversity in Holoenzyme Architecture and Extensive Conformational Variability in Biotin-Dependent Carboxylases, Advances in Protein Chemistry and Structural Biology, vol. 109, Elsevier, pp. 161–194, doi:10.1016/bs.apcsb.2017.04.006, ISBN 978-0-12-811876-4
- ↑ Charles O. Rock, Suzanne Jackowski, John E. Cronan, Chapter 2 - Lipid metabolism in prokaryotes, Editor(s): Dennis E. Vance, Jean E. Vance, New Comprehensive Biochemistry, Elsevier, Volume 31, 1996, Pages 35-74, ISSN 0167-7306, ISBN 978-0-444-82359-5, doi:10.1016/S0167-7306(08)60509-8.
- ↑ Lennarz, William J., and M. Daniel Lane. Encyclopedia of Biological Chemistry. Elsevier, 2013.