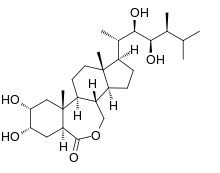
 | |
| Names | |
|---|---|
| IUPAC name
(22R,23R)-2α,3α,22,23-Tetrahydroxy-6,7-seco-5α-campestano-6,7-lactone | |
| Systematic IUPAC name
(1R,3aS,3bS,6aS,8S,9R,10aR,10bS,12aS)-1-[(2S,3R,4R,5S)-3,4-Dihydroxy-5,6-dimethylheptan-2-yl]-8,9-dihydroxy-10a,12a-dimethylhexadecahydro-6H-indeno[4,5-d][2]benzoxepin-6-one | |
| Other names
2,3,22,23-Tetrahydroxy-β-homo-7-oxaergostan-6-one | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChemSpider | |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C28H48O6 | |
| Molar mass | 480.686 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
Brassinolide is a plant hormone. The first isolated brassinosteroid, it was discovered when it was shown that pollen from rapeseed (Brassica napus) could promote stem elongation and cell division.[1] The biologically active component was isolated and named brassinolide.[2]
Biosynthesis
The production of brassinolide begins with a closely related sterol called campesterol, which is found in the cell membrane. Initially, it is reduced by an enzyme called DET2. This is followed by a series of oxidation reactions, facilitated by cytochrome P-450 enzymes, which add hydroxyl groups to the molecule. The most biologically significant of these reactions is the C6 oxidation, where a ketone is formed at the C6 carbon position. This single reaction increases the biological activity of the molecule by a factor of 200. Depending on when this C6 oxidation occurs, it is referred to as either the early or late C6 oxidation pathway. Both of these synthetic pathways have been observed in Arabidopsis seedlings. It appears that the late C6 oxidation pathway predominates when the seedlings are exposed to light, while the early pathway is active in the absence of light. If the plant cannot perform C6 oxidation, it results in the "Dwarf phenotype," characterized by severe growth deficits.[3]
Finally, in Arabidopsis, the Baeyer-Villiger lactonization process occurs through the action of the two homologous enzymes CYP85A1 and CYP85A2, leading to the formation of brassinolide.[4] Alternatively, there is a suggested synthetic pathway that starts from cholesterol, giving rise to C27 brassinosteroids.[5]
Mechanism of Action
Signal Transduction
Brassinosteroids, particularly the potent brassinolide, play a crucial role in controlling various plant processes such as germination, aging, and the ability to withstand environmental and biological stresses.[6] Because of this, researchers from around the world have extensively studied model organisms like Catharanthus roseus and Arabidopsis since they were first isolated in 1979.[7] These organisms have been thoroughly examined, from how they receive brassinolide signals to how these signals affect gene expression.
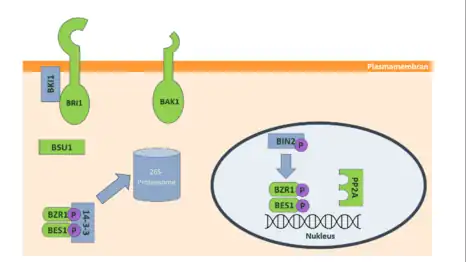
In Arabidopsis, the process begins with the BRI1 receptor (Brassinosteroid Insensitive 1 receptor). This receptor is a type of protein called a leucine-rich receptor kinase and allows brassinolide to attach to it from outside the cell. This binding causes a change in the receptor's shape, and it then interacts with another protein called BRI1 associated receptor kinase 1 (BAK1). This interaction results in both proteins being chemically modified by the addition of phosphate groups in a process called phosphorylation. This, in turn, sets off a chain reaction within the cell, activating some proteins and inhibiting others, including various kinases, phosphatases, and transcription factors. Among the activated proteins are the BR signaling kinases (BSK1, BSK2, and BSK3). Their activation, in turn, activates the phosphatase BRI1 suppressor1 (BSU1), which removes a phosphate group from another protein called brassinosteroid insensitive 2 (BIN2). Removing this phosphate group inactivates BIN2, an important enzyme. As a result, protein phosphotase 2A (PP2A) can remove phosphate groups from two transcription factors, brassinazole-resistant-1 (BZR1) and BRI1-EMS-suppressor-1 (BES1), allowing them to accumulate within the cell's nucleus. There, they control the expression of specific target genes, which are involved in various cellular processes. However, when there's no brassinolide around, a regulator called BRI1 kinase inhibitor (BKI1) prevents the BRI1 receptor from interacting with the BAK1 co-receptor. This prevents the activation of BIN2, causing BZR1 and BES1 to be chemically modified by adding phosphate groups. These modified transcription factors then interact with a protein called 14-3-3 and accumulate in the cell's cytoplasm. Eventually, they are broken down and degraded by a 26S proteasome. In this way, BIN2 kinase serves as an essential negative regulator, dampening the activity of the central transcription factors BES1 and BZR1.[6]
Homeostasis
The two transcription factors BES1 and BZR1 regulate a large number of genes involved in the synthesis of hormones, growth processes and stress response. In addition, there is inhibition of BR biosynthesis, which is evident as early as 15 minutes after brassinolide treatment.[8] Thus, the expression of several biosynthetic genes such as the CPD, DWF4 and CYP85A2 gene is inhibited.[6] These encode for brassinolide biosynthetic enzymes, thus the CPD gene encodes for the cytochrome P450 monooxygenase, which in the late C6 oxidation pathway oxidizes 6-deoxo-cathasterone to 6-deoxo teasterone and in the early C6 oxidation step catalyzes the reaction of cathasterone to teasterone. The enzyme CYP85A2 catalyzes the final biosynthesis step namely the Baeyer-Villiger lactonization of castasterone to brassinolide. This negative feedback loop ensures homeostasis of the hormone brassinolide.

Cell growth
Brassinolide induces genes that promote growth such as TCH4 and SAUR-Ac. The gene SAUR-Ac stands for small auxin upregulated RNAs, these belong to the auxin hormone induced genes and provide cell expansion.[9] Thus, SAURs inhibit the phosphatase PP2C-D so that the plasma membrane H+-ATPase is not dephosphorylated. The active phosphorylated proton pump can thus establish an electrochemical gradient in the cell wall. The acidity increases from 6 to 4.5-6 and according to the acid growth hypothesis, it ensures the activation of expansins that cleave the bond of cellulose and hemicellulose.[10] TCH4 was identified as xyloglucan endotransglycosylase (XETs) by sequence analysis and enzyme activity. Its main function is the modification of cell walls.[11] Thus, hemicellulose is composed of xyloglucans, which is built from 1,4-ß-linked glucose polymers with lateral 1,6-ß-linked xylene residues.[12] The xylogucans can form hydrogen bonds with the cellulose microfibrils and thus structurally stabilize the cell wall. This means XET can modify the cell wall structure. It cleaves xyloglucan molecules, stores some of the energy, and then consumes it again after expansion for linking. Thus, during cell migration expansion, XET can further loosen the cell wall, which provides for the absorption of water. The resulting internal pressure (turgor) is compensated for by the cell wall expansion, so that after re-linking the result is an expanded cell.[11]
Cross talk with other phytohormones
Gibberellin
Brassinolide and gibberellin are both interdependent. BZR1 in Arabidsopsis inhibits DELLAs a negative regulator of gibberellin transduction. Thus, the activity at the promoter for binding is reduced, so gibberellin promotes cell elongation. And DELLAs interaction provides a decrease in BZR1 binding affinity to DNA.
Jasmonic acid
Brassinolide inhibits jasmonic acid (JA) induced pathogen defensive for an ideal trade-off between growth and defense. Here, the transcription factor BES1 inhibits gene expression of PDF1.2a and PDF1.2b to reduce that of antimicrobial protein defensins. Furthermore, BES1 interacts with the transcription factors MYBs and reduces their activity to reduce glucosinolate (GS) biosynthesis, which is an important precursor for defense substances against predators.[13]
References
- ↑ Mitchell JW, Mandava N, Worley JF, Plimmer JR, Smith MV (1970). "Brassins--a new family of plant hormones from rape pollen". Nature. 225 (5237): 1065–6. Bibcode:1970Natur.225.1065M. doi:10.1038/2251065a0. PMID 16056912. S2CID 4116426.
- ↑ Grove MD, Spencer GF, Rohwedder WK, Mandava N, Worley JF, Warthen JD, Steffens GL, Flippen-Anderson JL, Cook JC (1979). "Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen". Nature. 281 (5728): 216–217. Bibcode:1979Natur.281..216G. doi:10.1038/281216a0. S2CID 4335601.
- ↑ Gerard J. Bishop, Takahito Nomura, Takao Yokota, Kate Harrison, Takahiro Noguchi, Shozo Fujioka, Suguru Takatsuto, Jonathan D. G. Jones, Yuji Kamiya (1999), "The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis", Proceedings of the National Academy of Sciences, vol. 96, no. 4, pp. 1761–1766, Bibcode:1999PNAS...96.1761B, doi:10.1073/pnas.96.4.1761, PMC 15587, PMID 9990098
{{citation}}: CS1 maint: multiple names: authors list (link) - ↑ Tae-Wuk Kim ,Jung-Yun Hwang, Young-Soo Kim, Se-Hwan Joo, Soo Chul Chang, June Seung Lee, Suguru Takatsuto, Seopng-Ki Kim (2005), "Arabidopsis CYP85A2, a Cytochrom P450, mediates the Baeyer-Villiger Oxidation of Castasterone to Brassinolide in Brassinosteroid Biosynthesis", The Plant Cell, vol. 17, no. 8, pp. 2397–2412, doi:10.1105/tpc.105.033738, PMC 1182497, PMID 16024588
{{citation}}: CS1 maint: multiple names: authors list (link) - ↑ Tae-Wuk Kim, Soo Chul Chang, June Seung Lee, Suguru Takatsuto, Takao Yokota, Seong-Ki Kim (2004), "Novel Biosynthetic Pathway of Castasterone from Cholesterol in Tomato", Plant Physiology, vol. 135, no. 3, pp. 1231–1242, doi:10.1104/pp.104.043588, PMC 519043, PMID 15247383
{{citation}}: CS1 maint: multiple names: authors list (link) - 1 2 3 Cécile Vriet, Eugenia Russinova, Christophe Reuzeau (2013), "From Squalene to Brassinolide: The Steroid Metabolic and Signaling Pathways across the Plant Kingdom", Molecular Plant, vol. 6, no. 6, pp. 1738–1757, doi:10.1093/mp/sst096, PMID 23761349
{{citation}}: CS1 maint: multiple names: authors list (link) - ↑ Gerald J. Bishop, Csaba Koncz (2002), "Brassinosteroids and Plant Steroid Hormone Signaling", The Plant Cell, vol. 14, no. Suppl, pp. S97–S110, doi:10.1105/tpc.001461, PMC 151250, PMID 12045272
- ↑ Jun-Xian He, Joshua M. Gendron, Yu Sun, Srinivas S. L. Gampala, Nathan Gendron, Catherine Qing Sun, Zhi-Yong Wang (2005), "BZR1 Is a Transcriptional Repressor with Dual Roles in Brassinosteroid Homeostasis and Growth Responses", Science, vol. 307, no. 5715, pp. 1634–1638, Bibcode:2005Sci...307.1634H, doi:10.1126/science.1107580, PMC 2925132, PMID 15681342
{{citation}}: CS1 maint: multiple names: authors list (link) - ↑ Angela K. Spartz, Hong Ren, Mee Yeon Park, Kristin N. Grandt, Sang Ho Lee, Angus S. Murphy, Michael R. Sussman, Paul J. Overvoorde, William M. Gray (2014), "SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis", The Plant Cell, vol. 26, no. 5, pp. 2129–2142, doi:10.1105/tpc.114.126037, PMC 4079373, PMID 24858935
{{citation}}: CS1 maint: multiple names: authors list (link) - ↑ Pyae Phyo, Ying Gu, Mei Hong (2019), "Impact of acidic pH on plant cell wall polysaccharide structure and dynamics: insights into the mechanism of acid growth in plants from solid-state NMR", Cellulose, vol. 26, no. 1, pp. 291–304, doi:10.1007/s10570-018-2094-7, hdl:1721.1/131772, S2CID 106390488
{{citation}}: CS1 maint: multiple names: authors list (link) - 1 2 Wei Xu, Mary M. Purugganan, Diana H. Polisensky, Danuta M. Antosiewicz, Stephen C. Fry, Janet Braam (1995), "Arabidopsis TCH4, Regulated by Hormones and the Environment, Encodes a Xyloglucan Endotransglycosylase", The Plant Cell, vol. 7, no. 10, pp. 1555–1567, doi:10.2307/3870019, JSTOR 3870019
{{citation}}: CS1 maint: multiple names: authors list (link) - ↑ M. M. Purugganan, J. Braam, S. C. Fry (1997), "The Arabidopsis TCH4 Xyloglucan Endotransglycosylase (Substrate Specificity, pH Optimum, and Cold Tolerance)", Plant Physiology, vol. 115, no. 1, pp. 181–190, doi:10.1104/pp.115.1.181, PMC 158473, PMID 9306698
{{citation}}: CS1 maint: multiple names: authors list (link) - ↑ Alfredo Kono, Yanhai Yin (2020), "Updates on BES1/BZR1 Regulatory Networks Coordinating Plant Growth and Stress Responses", Frontiers in Plant Science, vol. 11, doi:10.3389/fpls.2020.617162, PMC 7744466, PMID 33343611