The CARD-CC protein family is defined by an evolutionary conserved "caspase activation and recruitment domain" (CARD) and a coiled-coil (CC) domain.[1][2] Coiled-coils (CC) act as oligomerization domains for many proteins such as structural and motor proteins, and transcription factors. This means that monomers are converted to macromolecular complexes by polymerization.[3] In humans and other jawed vertebrates, the family consists of CARD9 and the three "CARD-containing MAGUK protein" (CARMA)[4] proteins CARD11 (CARMA1), CARD14 (CARMA2) and CARD10 (CARMA3). Although the MAGUK protein DLG5 contains both a CARD domain and a CC domain, it does not belong to the same family as the CARD-CC proteins since the evolutionary origin of its CARD domain is very likely to be different.[5]

Evolution and species distribution
The protein family is ancient and can be found as far back as Cnidaria, but has almost exclusively been studied in humans and mice. Notably, the protein family is absent in insects and nematodes, which makes it impossible to study its function in the most popular invertebrate model organisms (Drosophila and C. elegans). Invertebrates only have a CARD9-like ancestral CARD-CC member, and the earliest occurrence of a CARD-CC member with the CARMA domain composition is in the jawless vertebrate hagfish. Already in sharks are all four CARD-CC family members present, indicating that the 3 distinct CARMA CARD-CC family members were formed by two duplication events just before or very early in the jawed vertebrate evolution, almost half a billion years ago. The four CARD-CC ohnologous members in mice and humans differ in expression domains, where CARD9 is mostly expressed in myelocytes, CARD11 in lymphocytes, while CARD10 and CARD14 are mostly expressed in non-haematopoetic cells. This gene expression differentiation between the four CARD-CC family members conserved at least as far back as frogs (Xenopus tropicalis) and fish (Danio rerio),[6] indicating that the four CARD-CC family members have had distinct functions since early jawed vertebrate evolution.
Functions
A common theme for all four CARD-CC family proteins in mice and humans is that they are activated by different protein kinase C isoforms,[7] and recruit BCL10 and the paracaspase MALT1 upon activation, forming a so-called CBM complex. There are four different CBM complexes, defined by which CARD-CC family member that is responsible for its assembly: CBM-9 (CARD9), CBM-1 (CARD11/CARMA1), CBM-2 (CARD14/CARMA2) and CBM-3 (CARD10/CARMA3).[8] CBM complex assembly results in recruitment of TRAF6 to MALT1 and downstream activation of NF-κB transcriptional activity and expression of pro-inflammatory cytokines. The different CARD-CC family members show different expression pattern and gain- or loss of function mutation in the different CARD-CC family proteins cause different phenotypes.
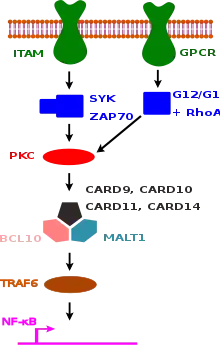
- Loss-of-function mutations in CARD9 disrupts lectin receptor signaling like Dectin 1, which causes enhanced susceptibility to fungal infections.[9][10]
- Gain-of-function variants in CARD9 appear to be associated to fibromyalgia, and possibly the comorbidities constipation, Irritable bowel syndrome and other psychosomatic sequelæ in the UK biobank association between exome sequences and health data.[11]
- Strong loss-of-function mutations in CARD11 cause severe defects in lymphocyte function since it is a critical downstream signal mediator in T- and B-cell antigen receptor signaling, which results in severe combined immunodeficiency (SCID).[12]
- Weak (hypomorphic) mutations in CARD11 causes atopic dermatitis disease.[13]
- Gain-of-function mutations in CARD11 can result in B-cell lymphoma or BENTA.[14][15]
- Gain-of-function mutations in CARD14 results in psoriasis or PRP.[16][17]
- There are indications that loss-of-function mutations in CARD14 could result in atopic dermatitis due to disrupted immune responses against skin microbes.[18]
- There are no obvious gain- or loss-of-function mutations in CARD10, but it is sometimes over-expressed in certain cancer variants,[19] and there is a SNP in CARD10 associated to glaucoma.[20]
References
- ↑ Staal J, Driege Y, Haegman M, Borghi A, Hulpiau P, Lievens L, et al. (2018). "Ancient Origin of the CARD-Coiled Coil/Bcl10/MALT1-Like Paracaspase Signaling Complex Indicates Unknown Critical Functions". Frontiers in Immunology. 9: 1136. doi:10.3389/fimmu.2018.01136. PMC 5978004. PMID 29881386.
- ↑ Bouchier-Hayes, Lisa; Martin, Seamus J (July 2002). "CARD games in apoptosis and immunity". EMBO Reports. 3 (7): 616–621. doi:10.1093/embo-reports/kvf139. ISSN 1469-221X. PMC 1084193. PMID 12101092. Retrieved 2023-09-11.
- ↑ "CC Protein Domain | Coiled Coil | Cell Signaling Technology". www.cellsignal.com. Retrieved 2020-02-01.
- ↑ Scudiero I, Vito P, Stilo R (August 2014). "The three CARMA sisters: so different, so similar: a portrait of the three CARMA proteins and their involvement in human disorders". Journal of Cellular Physiology. 229 (8): 990–7. doi:10.1002/jcp.24543. PMID 24375035. S2CID 45905503.
- ↑ Friedrichs, Frauke; Henckaerts, Liesbet; Vermeire, Severine; Kucharzik, Torsten; Seehafer, Tanja; Möller-Krull, Maren; Bornberg-Bauer, Erich; Stoll, Monika; Weiner, January (2008-04-01). "The Crohn's disease susceptibility gene DLG5 as a member of the CARD interaction network". Journal of Molecular Medicine. 86 (4): 423–432. doi:10.1007/s00109-008-0307-5. ISSN 1432-1440. PMID 18335190. S2CID 24982374. Retrieved 2023-09-11.
- ↑ "BioGPS". BioGPS. The Scripps Research Institute. Retrieved 2021-07-27.
- ↑ Staal, Jens; Driege, Yasmine; Haegman, Mira; Kreike, Marja; Iliaki, Styliani; Vanneste, Domien; Lork, Marie; Afonina, Inna S.; Braun, Harald; Beyaert, Rudi (2020-08-13). "Defining the combinatorial space of PKC::CARD-CC signal transduction nodes". The FEBS Journal. 288 (5): 1630–1647. doi:10.1111/febs.15522. ISSN 1742-4658. PMID 32790937. S2CID 221123226.
- ↑ Gehring, Torben; Seeholzer, Thomas; Krappmann, Daniel (2018). "BCL10 – Bridging CARDs to Immune Activation". Frontiers in Immunology. 9: 1539. doi:10.3389/fimmu.2018.01539. ISSN 1664-3224. PMC 6039553. PMID 30022982.
- ↑ Gross O, Gewies A, Finger K, Schäfer M, Sparwasser T, Peschel C, et al. (August 2006). "Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity". Nature. 442 (7103): 651–6. Bibcode:2006Natur.442..651G. doi:10.1038/nature04926. PMID 16862125. S2CID 4405404.
- ↑ Online Mendelian Inheritance in Man (OMIM): 607212
- ↑ Jens Staal; Yasmine Driege; Femke Van Gaever; Jill Steels; Rudi Beyaert (2023-01-01). ""Franken-CARD9": chimeric proteins for high-resolution studies of activating mutations in CARD10, CARD11 and CARD14". bioRxiv: 2023–03.06.531260. doi:10.1101/2023.03.06.531260. S2CID 257431611.
- ↑ Stepensky P, Keller B, Buchta M, Kienzler AK, Elpeleg O, Somech R, et al. (February 2013). "Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects" (PDF). The Journal of Allergy and Clinical Immunology. 131 (2): 477–85.e1. doi:10.1016/j.jaci.2012.11.050. hdl:1885/11324. PMID 23374270.
- ↑ Ma CA, Stinson JR, Zhang Y, Abbott JK, Weinreich MA, Hauk PJ, et al. (August 2017). "Germline hypomorphic CARD11 mutations in severe atopic disease". Nature Genetics. 49 (8): 1192–1201. doi:10.1038/ng.3898. PMC 5664152. PMID 28628108.
- ↑ Brohl AS, Stinson JR, Su HC, Badgett T, Jennings CD, Sukumar G, et al. (January 2015). "Germline CARD11 Mutation in a Patient with Severe Congenital B Cell Lymphocytosis". Journal of Clinical Immunology. 35 (1): 32–46. doi:10.1007/s10875-014-0106-4. PMC 4466218. PMID 25352053.
- ↑ Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, et al. (March 2008). "Oncogenic CARD11 mutations in human diffuse large B cell lymphoma". Science. 319 (5870): 1676–9. Bibcode:2008Sci...319.1676L. doi:10.1126/science.1153629. PMID 18323416. S2CID 26344383.
- ↑ Jordan CT, Cao L, Roberson ED, Pierson KC, Yang CF, Joyce CE, et al. (May 2012). "PSORS2 is due to mutations in CARD14". American Journal of Human Genetics. 90 (5): 784–95. doi:10.1016/j.ajhg.2012.03.012. PMC 3376640. PMID 22521418.
- ↑ Online Mendelian Inheritance in Man (OMIM): 607211
- ↑ Peled A, Sarig O, Sun G, Samuelov L, Ma CA, Zhang Y, et al. (January 2019). "Loss-of-function mutations in caspase recruitment domain-containing protein 14 (CARD14) are associated with a severe variant of atopic dermatitis". The Journal of Allergy and Clinical Immunology. 143 (1): 173–181.e10. doi:10.1016/j.jaci.2018.09.002. PMID 30248356.
- ↑ McAuley JR, Freeman TJ, Ekambaram P, Lucas PC, McAllister-Lucas LM (2018). "CARMA3 Is a Critical Mediator of G Protein-Coupled Receptor and Receptor Tyrosine Kinase-Driven Solid Tumor Pathogenesis". Frontiers in Immunology. 9: 1887. doi:10.3389/fimmu.2018.01887. PMC 6104486. PMID 30158935.
- ↑ Zhou T, Souzeau E, Sharma S, Siggs OM, Goldberg I, Healey PR, et al. (November 2016). "CARD10 enriched in primary open-angle glaucoma". Molecular Genetics & Genomic Medicine. 4 (6): 624–633. doi:10.1002/mgg3.248. PMC 5118207. PMID 27896285.