| Death effector domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
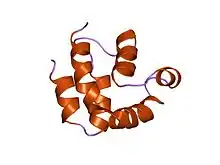 structure of the FADD (Mort1) death-effector domain.[1] | |||||||||||
| Identifiers | |||||||||||
| Symbol | DED | ||||||||||
| Pfam | PF01335 | ||||||||||
| InterPro | IPR001875 | ||||||||||
| SMART | DED | ||||||||||
| PROSITE | PS50168 | ||||||||||
| SCOP2 | 1a1z / SCOPe / SUPFAM | ||||||||||
| CDD | cd00045 | ||||||||||
| |||||||||||
The death-effector domain (DED) is a protein interaction domain found only in eukaryotes that regulates a variety of cellular signalling pathways.[2] The DED domain is found in inactive procaspases (cysteine proteases) and proteins that regulate caspase activation in the apoptosis cascade such as FAS-associating death domain-containing protein (FADD). FADD recruits procaspase 8 and procaspase 10 into a death induced signaling complex (DISC). This recruitment is mediated by a homotypic interaction between the procaspase DED and a second DED that is death effector domain in an adaptor protein that is directly associated with activated TNF receptors. Complex formation allows proteolytic activation of procaspase into the active caspase form which results in the initiation of apoptosis (cell death). Structurally the DED domain are a subclass of protein motif known as the death fold and contains 6 alpha helices, that closely resemble the structure of the Death domain (DD).
Structure
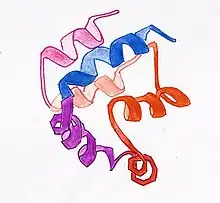
DED is a subfamily of the DD superfamily (other recognizable domains in this superfamily are: caspase-recruitment domain (CARD), pyrin domain (PYD) and death domain (DD)). The subfamilies resemble structurally one another, all of them (and DED in particular) are composed of a bundle of 6 alpha-helices, but they diverge in the surface features.
The complete primary structure of this proteic domain has not been consensually defined. Some studies described residues 2-184, but C-terminus and N-terminus residues are not identified yet. The presence of amino acids that determine the solubility and aggregation to DED allowed to identify DED's in different proteins, such as caspase-8 and MC159. The secondary structure of the domain, as said, is built by 6 alpha-helices.
The tertiary structure of the domain has been described from the crystallization of caspase 8 in the human. The method used to describe the structure was X-RAY diffraction and the resolution obtained is 2.2 Å.[3] DEDs in this protein show an asymmetric unit dimer, with its interface contains two hydrogen bonding networks, that appear as a filamentous structure. DED's function is determined by its structure. As far as it is known, the homotypic interactions that activate caspase and trigger apoptosis are mediated by asymmetrical surface contacts between partners (like DED1 and DED2 in the caspase-8 case).[4] The residues that form the surfaces are typically charged amino acids, but a short hydrophobic patch can also be observed on the interactive surface of the domain.
Function
DED domain is best known for its role in apoptosis. However, DED-containing proteins are also involved in other cellular processes so that they control both life and death cell decisions.
Extrinsic apoptosis
[5] Apoptosis is a controlled and programmed cell death that confers advantages during an organism lifecycle. The extrinsic pathway is directed by a family of proteases which become active in response to death stimuli. To know the role of DEDs in this process is important to observe the formation of the multiprotein death-including signalling complex (DISC).
DR4, TRAIL-R2 and CD95 are death receptors (members of TNF receptor superfamily) which interact together using their intracellular death domains (DDs). The DD of FADD, a protein containing a DED, can then interact with these DDs described. Here the function of FADD DED is to create a stabilized structure by self-associating FADD.

[6] These interactions are defined by helices α1/α4 and α2/α3: residues Ser1, Val6, His9, Leu43, Asp44 and Glu51 from α1/α4 are in contact with Thr21, Phe25, Lys33, Arg34, Glu37 and Glu51 from α2/α3 of the second molecule. Each interaction involves an area of 1062 Å2 and contributions from hydrophobic side chains, hydrogen bonding and salt bridges. The final homodimer has a structure oriented so that each subunit has the 2 interaction sites.
Procaspase-8, also a DED-containing protein, has affinity for the FADD DED. It's for that reason that they are recruited to FADD as monomers via their DEDs. These interaction is defined by α1/α4 of procapase-8 DED-A and FADD DED α2/α3 or α1/α4 of FADD DED and α2/α5 of procapase-8 DED-B. Procaspase-8 DED-B interacts with FADD and DED-A mediates capase-8 chain formation, or vice versa. However, in both cases the interaction leads to create a dimer between procaspases, which generates a conformational change. This dimerization is essential to create the active site; a p12 subunit is liberated and it is subsequently processed to the small p10 subunit. The two molecules of procapase-8 are associated with these p10 subunits creating an active protease-8 cell death.[7][8]
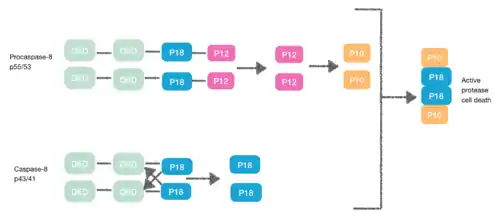
Necroptosis
[9] During the creation of the DISC procaspase-8 can also heterodimerise with another DED-containing protein known as FLIPL. FLIPL’s pseudo-caspase has two tandem DEDs that are very similar to the N-terminus of capase-8, but in which there is an important mutation in the active site (cysteine to tyrosine).
This heterodimeration done between their DEDs prevents from the normal homodimeration so that the pseudo-caspase is unable to activate the apoptotic cascade. FLIPL ’s pseudo-caspase is more efficient at inducing the conformational change. However, FLIPL hasn’t enough enzymatic activity so that cleavage between the DEDs and p18 it’s not possible. In consequence it’s impossible to create the active protease cell death.[10]
Procaspase-8 can also heterodimerise with FLIPS, also a DED containing protein. In this case heterodimerisation directly fails to activate procaspase-8 as the initial conformational change cannot take place in procaspase-8’s caspase domain.[10]
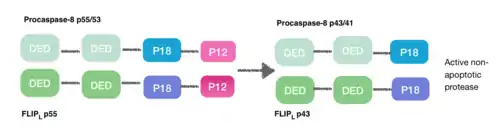
This is how DED can also inhibit the apoptosis cascade, and the consequence is necroptosis.
The DED protein family
DED-containing proteins
Caspase-8 and caspase-10
[11] Caspases are cysteine proteases responsible for dismantling off the cell during apoptosis.
These proteins are zymogens and become active after their cleavage at specific sites within the molecule.
Structure:
- Death Effector Domain (DED) and a Caspase Recruitment Domain (CARD) that are englobed in a structure called pro-domain, which is located at the N-terminus
- Catalytic protease domain at the C-terminus.
There are two groups of proteases:
- Effector caspases: induce the biggest part of the morphological changes that occur during apoptosis.
- Initiator caspases: responsible for the activation of effector caspases. These caspases are activated through oligomerization and cleavage that make the protein functional.
The two tandem DEDs in the pro-domain of caspase induce the protein-protein interactions with other proteins like the FADD.
Studying caspases is important since they don’t only control apoptosis but also inhibit it, depending on the necessity of the cell. Scientists find that they are a mechanism that can regulate cell life and is important for cancer therapies.
FLICE-like inhibitory proteins (FLIPs)
FLICE-like inhibitory proteins (FLIPs) are cell inhibitors capable of stopping the death receptors’ signal, which cause cell apoptosis.
The first FLIPs that were identified were expressed by γ-herpes viruses so they were called v-FLIPs. These v-FLIPs were able to associate with the receptor in the death-inducing signaling complex (DISC), blocking that way the CD95-mediated apoptosis.
[12]vFLIPs predominantly contain two sequential DEDs, which are highly homologous to the N-terminus of caspase-8.
[10] The cellular homologues of v-FLIPs are generally expressed in two forms:
- c-FLIPS (short): it contains only the amino-terminal tandem DEDs followed by a short carboxy-terminal section. Its structure is similar to the viral FLIPs.
- c-FLIPL (long): it consists of not only the tandem DEDs, but also a protease-like domain (homologous to caspase-8) in which different important for protease activity amino acids are mutated, including the active site cysteine.
[12] Both forms of c-FLIP are draft to the CD95 DISC, where they heterodimerize with caspase-8. c-FLIP has been involved in signaling alternative pathways, connecting the CD95 receptor to the NF-κB, JNK and MAPK pathways.
PEA-15/PED
PEA-15 (Phosphoprotein Enriched in Astrocytes-15 kDa) also known as PED (Phosphoprotein Enriched in Diabetes) is a DED-containing protein with pleiotropic effects.
PED is a small, non-catalytic, protein consisting of an N-terminal death-effector domain (DED) and a C-terminal tail with irregular structure.[13] PED/PEA-15 interacts with various types of proteins with and without DEDs, and its specificity of joining these proteins is mediated by the phosphorylation on two serine residues on the C-terminal tail:
- Ser104: phosphorylated by protein kinase C (PKC).
- Ser116: the substrate for calcium/calmodulin-dependent protein kinase II (CamKII).
[13] PEA-15 works as an antiapoptotic DED protein in several signaling cascades. In TNF α-, CD95- and TRAIL-mediated pathways, PEA-15 acts binding and disrupting FADD and caspase-8 interactions.
[10] Besides apoptosis, PEA-15 inhibits the insulin-mediated glucose transport in muscle cells, so a high level expression of PEA-15's mRNA has been associated to diabetes mellitus type II.
DEDD/DEDD2
Death effector domain containing DNA binding (DEDD). Shows DNA binding capacity, localized in the nucleoli in overexpression where it associates with a molecule called DEDAF (DED-associated factor) that potentiates apoptosis. In addition it blocks RNA polymerase I transcription by binding to the DNA.
DEDD2 (FLAME-3) is a DEDD homologue that shares a 48.5% of the amino acidic sequence. It is noted to interact with c-FLIP and DEDD and to have an important role in polymerase II-dependent transcription repression.
Proteins with a DED-related domain
HIP-1 and HIPPI
Huntingtin interacting protein-1 (HIP-1) is a protein that interacts with huntingtin (Htt), another protein that when is mutated (with expanded polyglutamine repeats) forms protein aggregates in the brain of patients with Huntington's disease (HD).
[14] HIP-1 contains a pseudo death effector domain (pDED), that's why the overexpression of HIP-1 induces apoptosis in several cells as DED proteins do. This type of apoptosis depends on the pDED of the HIP-1, and it consists in the activation of caspase-3, an enzyme that is reduced when wild-type Htt is expressed, that fact suggests that HIP-1 cooperates with Htt in the pathomechanism of Huntington's disease.
[10] By yeast two-hybrid screening, HIP-1 has shown to interact with a protein of 419 amino acids called HIPPI (HIP-1 protein interactor). Succeeding experiments have revealed that the presence of HIPPI determines the HIP-1-induced apoptosis.
FLASH
FLICE-associated huge protein. Contains a similar domain to DED, but the homology is very weak and its function is still unclear.
Therapeutically exploiting DED
[7] DED complexes have been shown to function at crucial steps controlling life and death cell processes. This knowledge is particularly useful in therapy because there are so many pathologies related with an abnormal control of the cell life.
The absence of apoptosis is feature of cancer. In some cases the gene encoding procaspase-8 is silenced by methylation, so it is necessary to activate the gene using epigenetic treatments to have active protease. In other cases there is an overexpression of FLIP, the anti-apoptotic molecule that prevents from the formation of the active caspase. In this case there are some anti-cancer agents that downregulate FLIP expression.
However, the abnormal apoptosis it is not exclusive from cancer, there are other pathologies such as inflammation and neurodegenerative diseases than can also be treated with these kind of therapeutics.
References
- ↑ Eberstadt M, Huang B, Chen Z, et al. (April 1998). "NMR structure and mutagenesis of the FADD (Mort1) death-effector domain". Nature. 392 (6679): 941–5. doi:10.1038/31972. PMID 9582077. S2CID 4370202.
- ↑ Valmiki MG, Ramos JW (March 2009). "Death effector domain-containing proteins". Cell. Mol. Life Sci. 66 (5): 814–30. doi:10.1007/s00018-008-8489-0. PMID 18989622. S2CID 13117680.
- ↑ Shen, Chen; Yue, Hong; Pei, Jianwen; Guo, Xiaomin; Wang, Tao; Quan, Jun-Min (2015). "Crystal structure of the death effector domains of caspase-8". Biochemical and Biophysical Research Communications. 463 (3): 297–302. doi:10.1016/j.bbrc.2015.05.054. ISSN 0006-291X. PMID 26003730.
- ↑ "Structures, Domains and Function in Cell Death".
- ↑ Elmore, Susan (2007). "Apoptosis: A Review of Programmed Cell Death". Toxicologic Pathology. 35 (4): 495–516. doi:10.1080/01926230701320337. ISSN 1533-1601. PMC 2117903. PMID 17562483.
- ↑ Singh, Nitu; Hassan, Ali; Bose, Kakoli (2015). "Molecular basis of death effector domain chain assembly and its role in caspase-8 activation". The FASEB Journal. 30 (1): 186–200. doi:10.1096/fj.15-272997. ISSN 1530-6860. PMID 26370846.
- 1 2 Riley, JS; Malik, A; Holohan, C; Longley, DB (2015). "DED or alive: assembly and regulation of the death effector domain complexes". Cell Death and Disease. 6 (8): e1866. doi:10.1038/cddis.2015.213. ISSN 2041-4889. PMC 4558505. PMID 26313917.
- ↑ Yao, Zhan; Duan, Shanshan; Hou, Dezhi; Heese, Klaus; Wu, Mian (2007). "Death effector domain DEDa, a self-cleaved product of caspase-8/Mch5, translocates to the nucleus by binding to ERK1/2 and upregulates procaspase-8 expression via a p53-dependent mechanism". The EMBO Journal. 26 (4): 1068–1080. doi:10.1038/sj.emboj.7601571. ISSN 1460-2075. PMC 1852837. PMID 17290218.
- ↑ Lee, Eun-Woo; Seo, Jinho; Jeong, Manhyung; Lee, Sangsik; Song, Jaewhan (2012). "The roles of FADD in extrinsic apoptosis and necroptosis". BMB Reports. 45 (9): 496–508. doi:10.5483/BMBRep.2012.45.9.186. ISSN 1976-670X. PMID 23010170.
- 1 2 3 4 5 Barnhart, Bryan C; Lee, Justine C; Alappat, Elizabeth C; Peter, Marcus E (2003). "The death effector domain protein family". Oncogene. 22 (53): 8634–8644. doi:10.1038/sj.onc.1207103. ISSN 0950-9232. PMID 14634625.
- ↑ Schleich, K.; Buchbinder, J. H.; Pietkiewicz, S.; Kähne, T.; Warnken, U.; Öztürk, S.; Schnölzer, M.; Naumann, M.; Krammer, P. H. (2016-04-01). "Molecular architecture of the DED chains at the DISC: regulation of procaspase-8 activation by short DED proteins c-FLIP and procaspase-8 prodomain". Cell Death & Differentiation. 23 (4): 681–694. doi:10.1038/cdd.2015.137. ISSN 1350-9047. PMC 4986640. PMID 26494467.
- 1 2 Yu, JW; Shi, Y (2008). "FLIP and the death effector domain family". Oncogene. 27 (48): 6216–6227. doi:10.1038/onc.2008.299. ISSN 0950-9232. PMID 18931689.
- 1 2 Twomey, Edward C; Cordasco, Dana F; Wei, Yufeng (2012). "Profound conformational changes of PED/PEA-15 in ERK2 complex revealed by NMR backbone dynamics". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1824 (12): 1382–1393. doi:10.1016/j.bbapap.2012.07.001. ISSN 1570-9639. PMID 22820249.
- ↑ Bhattacharyya, Nitai P; Banerjee, Manisha; Majumder, Pritha (2008). "Huntington's disease: roles of huntingtin-interacting protein 1 (HIP-1) and its molecular partner HIPPI in the regulation of apoptosis and transcription". The FEBS Journal. 275 (17): 4271–4279. doi:10.1111/j.1742-4658.2008.06563.x. ISSN 1742-464X. PMID 18637945.