Extrachromosomal circular DNA (eccDNA) is a type of double-stranded circular DNA structure that was first discovered in 1964 by Alix Bassel and Yasuo Hotta.[1] In contrast to previously identified circular DNA structures (e.g., bacterial plasmids, mitochondrial DNA, circular bacterial chromosomes, or chloroplast DNA), eccDNA are circular DNA found in the eukaryotic nuclei of plant and animal (including human) cells. Extrachromosomal circular DNA is derived from chromosomal DNA, can range in size from 50 base pairs to several mega-base pairs in length, and can encode regulatory elements and full-length genes. eccDNA has been observed in various eukaryotic species[2][3][4][5][6][7][8] and it is proposed to be a byproduct of programmed DNA recombination events, such as V(D)J recombination.[8][9]
Historical Background
In 1964, Bassel and Hotta published their initial discovery of eccDNA that they made while researching Franklin Stahl’s chromosomal theory.[10] In their experiments, they visualized isolated wheat nuclei and boar sperm by using electron microscopy.[10] Their research found that boar sperm cells contained eccDNA of various sizes.[10] In 1965, Arthur Spriggs’ research group identified eccDNA in the samples of five pediatric patients’ embryonic tumors and one adult patient’s bronchial carcinoma.[11] In the following years, additional research led to the discovery of eccDNA in various species listed in Table 1:
| Year | Organism | Reference |
|---|---|---|
| 1965 | Boar sperm | Hotta and Bassel, 1965[10] |
| 1965 | Human tumors | Cox et al., 1965[11] |
| 1969 | Yeast | Billheimer and Avers, 1969[12] |
| 1984 | Trypanosomatids | Beverly et al., 1984[13] |
| 1972 | Euglena | Nass and Ben-Shaul, 1972[14] |
| 1972 | Tobacco | Wong and Wildman, 1972[15] |
| 1972, 1978, 1980 | Fungi | Agsteribbe et al., 1972;[16] Stahl et al., 1978;[17] Lazarus et al., 1980[18] |
| 1972, 1985 | Cultured human fibroblasts | Smith and Vinograd, 1972;[19] Riabowol et al., 1985[4] |
| 1976 | Xenopus | Buongiorno-Nardelli et al., 1976[20] |
| 1978, 1984 | Chicken bursa | DeLap and Rush, 1978;[21] Toda and Yamagishi, 1984[22] |
| 1982 | Human tissues | Calabretta et al., 1982[23] |
| 1983 | Mouse embryo | Yamagishi et al., 1983[24] |
| 1983, 1988, 1990 | Mouse tissues | Tsuda et al., 1983;[25] Flores et al., 1988;[26] Gaubatz and Flores, 1990[2] |
| 1983 | Mouse thymocytes | Yamagishi et al., 1983[24] |
| 1983 | Mouse lymphocytes | Tsuda et al., 1983[26] |
21st Century Research
In the 21st century, researchers have focused on better characterizing the specific subtypes of eccDNA, as well as the structure and function of these molecules within biological systems:[27]
- In 2012, Shibata et al. discovered a specific type of eccDNA called microDNA.[6] The researchers found tens of thousands of microDNAs in mouse tissues and cell lines, as well as human cell lines.[6]
- In 2017, Turner et al. identified using whole-genome sequencing (WGS), cytogenetic analysis, and structural modeling that extrachromosomal circular DNA is highly amplified and common in various types of cancers.[28] They found that eccDNA molecules have significant heterogeneity between different cells even if they are derived from the same individual.[28] Furthermore, these eccDNA molecules contained tumor-driving genes and were reported to be rarely found in non-cancerous tissues.[28]
- In 2018, Møller et al. used healthy human muscle and blood cell samples to identify over 100,000 types of eccDNA, which suggested that eccDNA could be found within somatic cells ubiquitously.[29]
- In 2019, Wu et al. found that ecDNA (subtype of eccDNA) associates with chromatin, but unlike chromosomes it does not have higher-order compaction, which increases its accessibility.[30]
- In 2021, Wang et al. elaborated on the formation of eccDNAs and identified the immunostimulant function of eccDNAs.[31] They also developed an improved eccDNA purification protocol that decreases linear DNA contamination within purified samples.[31]
eccDNA Purification
Historically, eccDNA was purified using a two-step procedure that involved first isolating crude extrachromosomal DNA and subsequently digesting linear DNA via exonuclease digestion.[31] Yet, this technique often results in linear DNA contamination because exonuclease digestion is not sufficient to remove all linear DNA.[31] In 2021, Wang et al. developed a three-step eccDNA enrichment method that improved eccDNA purification:[31]
- The cells were first dehydrated in > 90% methanol. To extract crude extrachromosomal DNA, the cells were lysed with a pH 11.8 alkaline lysis buffer, neutralized with a neutralization buffer, and precipitated using a precipitation buffer. A commercial plasmid purification kit's silica column was used to isolate DNA from other cell components.
- The eluted DNA was digested with the restriction enzyme PacI to linearize mitochondrial DNA (mtDNA) and an exonuclease that can digest linear DNA.
- Finally, circular DNA was selectively recovered by a commercial solution and silica beads to remove linear DNA that was not removed by exonuclease digestion.
Double minutes (DM) vs. extrachromosomal circular DNA (eccDNA)
Initially, the term double minutes (DM) was commonly used to refer to extrachromosomal circular DNA because it often appeared as a pair in early studies.[27] As research has continued, different subtypes of extrachromosomal circular DNA have been identified that are not double minutes (e.g., microDNA). In 2014, Barreto et al. identified that double minutes only comprise roughly 30% of extrachromosomal DNA.[32] Thus, the term extrachromosomal circular DNA (eccDNA) is becoming more widely used, while the term double minutes is now reserved for a specific subtype of eccDNA.[32]
Structure
eccDNA are circular DNA that have been found in human, plant, and animal cells and are present in the cell nucleus in addition to the chromosomal DNA. eccDNA is distinguishable from other circular DNA in cells, such as mitochondrial DNA (mtDNA), because it ranges in size from a few hundred bases to megabases and is derived from genomic DNA.[1] For example, eccDNA can be formed from exons of protein coding genes, like mucin and titin. Researchers have hypothesized that eccDNA may contribute to the expression of different isoforms of a gene by interfering with or promoting the transcription of specific exons.[1]
eccDNA has been classified as one of four different categories of circular DNA based on size and sequence, including small polydispersed circular DNA (spcDNA), telomeric circles (t-circles), microDNA (100-400 bp), and extrachromosomal DNA (ecDNA).[27] Each of these types has its own unique biological characteristics (see Table 2):[27]
| Name of eccDNA | Size | Characteristics | Function |
|---|---|---|---|
| spcDNA | 100–10 kb | Highly diverse type of eccDNA, there is a large range of the number of spcDNA found cells | Involved in human genetic instability |
| Telomeric circles | multiples of 738 bp | Formed by telomeric arrays, which is a series of repeated sequences at the end of linear DNA. | Involved in the alternative lengthening of telomeres (ALT) |
| microDNA | 100-400 bp | Derived from genomic locations that have a high GC content and exon density | Express small functional regulatory RNAs (e.g., microRNAs and new is-like RNAs). |
| ecDNA | 1-3 Mb | Include full genes, no telomeres, acentric | Amplify genes involved in development of cancer and drug resistance |
eccDNA biogenesis
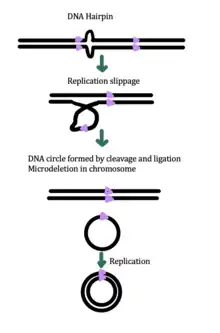
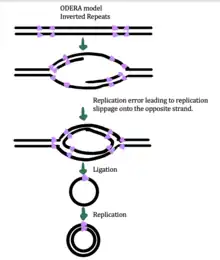

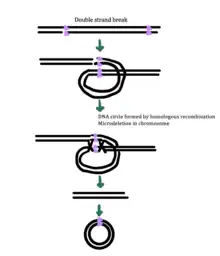
While the exact mechanism for eccDNA generation is still unknown, some studies have suggested that eccDNA generation might be linked to DNA damage repair,[33] hyper-transcription,[33][34] homologous recombination,[35] and replication stress.[33] There are multiple proposed mechanisms for eccDNA formation: (1) replication slippage creates a loop on the template strand that is then excised and ligated into a circle leaving a microdeletion on the chromosome, (2) replication slippage creates a loop in the product strand that is excised and ligated into a circle that does not generate a microdeletion in the chromosome, (3) the ODERA mechanism of eccDNA formation, and (4) a double stranded break in a repeat region is repaired by homologous recombination, during which the fragment forms a circle and the chromosome suffers a microdeletion[1]
Research conducted in 2021 demonstrated that apoptotic cells are a source of eccDNAs; this was concluded on account of the study showing that apoptotic DNA fragmentation (ADF) is a prerequisite for eccDNA formation through purification methods.[31]
eccDNA in non-cancerous cells
To test whether eccDNAs occur in non-cancer cells, mouse embryonic stem cells and Southern Blot analysis were used; the results confirmed that eccDNA is found in both cancerous and non-cancerous cells.[31] It is also known that eccDNA is unlikely to be derived from specific genome regions; sequencing data from 2021 reports that the data suggests eccDNAs are widespread across the entirety of the genome.[31] Genome mapping of full-length eccDNAs demonstrated their different genomic alignment patterns, which includes at adjacent, overlapped, or nested positions on the same chromosome or across different chromosomes.[31] eccDNAs originate mostly from single, continuous genomic loci, meaning that one single genomic fragment self-circularizes to form the eccDNA, rather than being formed from ligation of different genomic fragments.[31] These two variants can be classified as continuous and non-continuous eccDNAs, respectively.[31] To further understand the reason behind the circularization of fragmented DNA, the three various mammalian ligase enzymes were tested: Lig1, Lig3, and Lig4[31]. Using knockout models in the CH12F3 mouse B-lymphocyte cell line, research conducted in 2021 identified Lig3 as the main ligase for eccDNA generation in these cells.[31]
Function
The exact function of eccDNA has been debated, but some studies have suggested that eccDNAs might contribute to gene amplification in cancer,[1] immune function,[31] and aging.[34][35][36]
eccDNA function in immune system
According to research conducted in 2021, another function of eccDNAs is their role as possible immunostimulants.[31] eccDNA significantly induces type I interferons (IFNα, IFNβ), interleukin-6 (IL-6), and tumor necrosis factor (TNF), even more so than linear DNA and other generally potent cytokine inducers at their highest concentration levels.[31] Similar patterns are observed with macrophages as the data showed that eccDNAs are very potent immunostimulants in activating both bone marrow-derived dendritic cells and bone marrow-derived macrophages.[31] Additionally, experiments altered the eccDNA structure with one nick per eccDNA segment and subsequently treated with enzymes to generate linear versions of the eccDNA.[31] In these experiments, cytokine transcription, an important marker for immune system activity, was shown to be much higher in the non-treated eccDNA compared to the linearized treatment, conferring that the circular structure of eccDNA rather than the genetic sequence itself gives the eccDNA its immune function.[31]
eccDNA function in cancer
Some known functions of eccDNA include contributions to intercellular genetic heterogeneity in tumors, and more specifically the amplification of oncogenes and drug-resistant genes. This also supports that the genes on eccDNA are expressed. Overall, eccDNA has been linked to cancer and drug resistance, aging, gene compensation,[1] and for this reason it continues to be a significant topic of discussion.
Applications
Role in cancer
A subtype of eccDNA, such as ecDNA, ribosomal DNA locus (Extrachromosomal rDNA circle), and double minutes have been associated with genomic instability. Double minute ecDNAs are fragments of extrachromosomal DNA, which were originally observed in a large number of human tumors including breast, lung, ovary, colon, and most notably, neuroblastoma. They are a manifestation of gene amplification during the development of tumors, which give the cells selective advantages for growth and survival. Double minutes, like actual chromosomes, are composed of chromatin and replicate in the nucleus of the cell during cell division. Unlike typical chromosomes, they are composed of circular fragments of DNA, up to only a few million base pairs in size and contain no centromere or telomere.
Double minute chromosomes (DMs), which present as paired chromatin bodies under light microscopy, have been shown to be a subset of ecDNA.[28][37] Double minute chromosomes represent about 30% of the cancer-containing spectrum of ecDNA, including single bodies,[28] and have been found to contain identical gene content as single bodies. The ecDNA notation encompasses all forms of the large gene-containing extrachromosomal DNA found in cancer cells. This type of ecDNA is commonly seen in cancer cells of various histologies, but virtually never in normal tissue.[38][28] ecDNA are thought to be produced through double-strand breaks in chromosomes or over replication of DNA in an organism.[39]
The circular shape of ecDNA differs from the linear structure of chromosomal DNA in meaningful ways that influence cancer pathogenesis.[40][30] Oncogenes encoded on ecDNA have massive transcriptional output, ranking in the top 1% of genes in the entire transcriptome. In contrast to bacterial plasmids or mitochondrial DNA, ecDNA are chromatinized, containing high levels of active histone marks, but a paucity of repressive histone marks. The ecDNA chromatin architecture lacks the higher-order compaction that is present on chromosomal DNA and is among the most accessible DNA in the entire cancer genome.
From eccDNA, matrix attachment regions (MARs) were found to activate amplification of oncogenes.[1] Transfection of these MARs into human embryonic kidney 293T cells resulted in an increase in gene expression, suggesting that these eccDNA-derived MARs are involved in oncogene activation.[41] eccDNA also appears to play a role in other cancers such as breast cancer, where oncogenes in human epidermal growth factor receptor 2 (HER2)-positive breast cancer genes in eccDNA are amplified.[1] This eccDNA has also shown the ability to acquire resistance to therapies for receptor tyrosine kinases (RTKs), like HER26.[42]
Role in aging
Yeast are model organisms for studying aging, and eccDNAs have been shown to accumulate in old cells and play a role in causing aging in yeast.[36] Speculation continues on the generality of this concept in higher species, like mammals.[36]
See also
References
- 1 2 3 4 5 6 7 8 Paulsen T, Kumar P, Koseoglu MM, Dutta A (April 2018). "Discoveries of Extrachromosomal Circles of DNA in Normal and Tumor Cells". Trends in Genetics. 34 (4): 270–278. doi:10.1016/j.tig.2017.12.010. PMC 5881399. PMID 29329720.
- 1 2 3 Gaubatz JW (1990). "Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells". Mutation Research. 237 (5–6): 271–292. doi:10.1016/0921-8734(90)90009-g. PMID 2079966.
- ↑ Cohen S, Yacobi K, Segal D (June 2003). "Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila". Genome Research. 13 (6A): 1133–1145. doi:10.1101/gr.907603. PMC 403641. PMID 12799349.
- 1 2 Cohen S, Agmon N, Sobol O, Segal D (March 2010). "Extrachromosomal circles of satellite repeats and 5S ribosomal DNA in human cells". Mobile DNA. 1 (1): 11. doi:10.1186/1759-8753-1-11. PMC 3225859. PMID 20226008.
- ↑ Stanfield S, Helinski DR (October 1976). "Small circular DNA in Drosophila melanogaster". Cell. 9 (2): 333–345. doi:10.1016/0092-8674(76)90123-9. PMID 824055. S2CID 39382051.
- 1 2 3 Shibata Y, Kumar P, Layer R, Willcox S, Gagan JR, Griffith JD, Dutta A (April 2012). "Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues". Science. 336 (6077): 82–86. Bibcode:2012Sci...336...82S. doi:10.1126/science.1213307. PMC 3703515. PMID 22403181.
- ↑ Møller HD, Parsons L, Jørgensen TS, Botstein D, Regenberg B (June 2015). "Extrachromosomal circular DNA is common in yeast". Proceedings of the National Academy of Sciences of the United States of America. 112 (24): E3114–E3122. Bibcode:2015PNAS..112E3114M. doi:10.1073/pnas.1508825112. PMC 4475933. PMID 26038577.
- 1 2 Shoura MJ, Gabdank I, Hansen L, Merker J, Gotlib J, Levene SD, Fire AZ (October 2017). "Intricate and Cell Type-Specific Populations of Endogenous Circular DNA (eccDNA) in Caenorhabditis elegans and Homo sapiens". G3. 7 (10): 3295–3303. doi:10.1534/g3.117.300141. PMC 5633380. PMID 28801508.
- ↑ Hayday AC, Saito H, Gillies SD, Kranz DM, Tanigawa G, Eisen HN, Tonegawa S (February 1985). "Structure, organization, and somatic rearrangement of T cell gamma genes". Cell. 40 (2): 259–269. doi:10.1016/0092-8674(85)90140-0. PMID 3917858. S2CID 34582929.
- 1 2 3 4 Hotta Y, Bassel A (February 1965). "Molecular Size and Circularity of DNA in Cells of Mammals and Higher Plants". Proceedings of the National Academy of Sciences of the United States of America. 53 (2): 356–362. Bibcode:1965PNAS...53..356H. doi:10.1073/pnas.53.2.356. PMC 219520. PMID 14294069.
- 1 2 Cox D, Yuncken C, Spriggs AI (July 1965). "Minute Chromatin Bodies in Malignant Tumours of Childhood". Lancet. 1 (7402): 55–58. doi:10.1016/s0140-6736(65)90131-5. PMID 14304929.
- ↑ Billheimer FE, Avers CJ (October 1969). "Nuclear and mitochondrial DNA from wild-type and petite yeast: circularity, length, and buoyant density". Proceedings of the National Academy of Sciences of the United States of America. 64 (2): 739–746. Bibcode:1969PNAS...64..739B. doi:10.1073/pnas.64.2.739. PMC 223406. PMID 5261045.
- ↑ Beverley SM, Coderre JA, Santi DV, Schimke RT (September 1984). "Unstable DNA amplifications in methotrexate-resistant Leishmania consist of extrachromosomal circles which relocalize during stabilization". Cell. 38 (2): 431–439. doi:10.1016/0092-8674(84)90498-7. PMID 6467372. S2CID 2030494.
- ↑ Nass MM, Ben-Shaul Y (June 1972). "A novel closed circular duplex DNA in bleached mutant and green strains of Euglena gracilis". Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis. 272 (1): 130–136. doi:10.1016/0005-2787(72)90041-X. PMID 4625469.
- ↑ Wong FY, Wildman SG (January 1972). "Simple procedure for isolation of satellite DNA's from tobacco leaves in high yield and demonstration of minicircles". Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis. 259 (1): 5–12. doi:10.1016/0005-2787(72)90468-6. PMID 5011974.
- ↑ Agsteribbe E, Kroon AM, van Bruggen EF (May 1972). "Circular DNA from mitochondria of Neurospora crassa". Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis. 269 (2): 299–303. doi:10.1016/0005-2787(72)90439-X. PMID 4260513.
- ↑ Stahl U, Lemke PA, Tudzynski P, Kück U, Esser K (July 1978). "Evidence for plasmid like DNA in a filamentous fungus, the ascomycete Podospora anserina". Molecular & General Genetics. 162 (3): 341–343. doi:10.1007/BF00268860. PMID 683172. S2CID 12655440.
- ↑ Lazarus CM, Earl AJ, Turner G, Küntzel H (May 1980). "Amplification of a mitochondrial DNA sequence in the cytoplasmically inherited 'ragged' mutant of Aspergillus amstelodami". European Journal of Biochemistry. 106 (2): 633–641. doi:10.1111/j.1432-1033.1980.tb04611.x. PMID 6249580.
- ↑ Smith CA, Vinograd J (August 1972). "Small polydisperse circular DNA of HeLa cells". Journal of Molecular Biology. 69 (2): 163–178. doi:10.1016/0022-2836(72)90222-7. PMID 5070865.
- ↑ Buongiorno-Nardelli M, Amaldi F, Lava-Sanchez PA (March 1976). "Electron microscope analysis of amplifying ribosomal DNA from Xenopus laevis". Experimental Cell Research. 98 (1): 95–103. doi:10.1016/0014-4827(76)90467-5. PMID 1253845.
- ↑ DeLap RJ, Rush MG (December 1978). "Change in quantity and size distribution of small circular DNAs during development of chicken bursa". Proceedings of the National Academy of Sciences of the United States of America. 75 (12): 5855–5859. Bibcode:1978PNAS...75.5855D. doi:10.1073/pnas.75.12.5855. PMC 393074. PMID 282606.
- ↑ Toda M, Hirama T, Takeshita S, Yamagishi H (June 1989). "Excision products of immunoglobulin gene rearrangements". Immunology Letters. 21 (4): 311–316. doi:10.1016/0165-2478(89)90025-4. PMID 2767726.
- ↑ Calabretta B, Robberson DL, Barrera-Saldaña HA, Lambrou TP, Saunders GF (March 1982). "Genome instability in a region of human DNA enriched in Alu repeat sequences". Nature. 296 (5854): 219–225. Bibcode:1982Natur.296..219C. doi:10.1038/296219a0. PMID 6278320. S2CID 4265874.
- 1 2 Yamagishi H, Kunisada T, Iwakura Y, Nishimune Y, Ogiso Y, Matsushiro A (December 1983). "Emergence of the Extrachromosomal Circular DNA Complexes as One of the Earliest Signals of Cellular Differentiation in the Early Development of Mouse Embryo. (mouse embryo/teratocarcinoma/mica-press-adsorption/circular DNA complex/DNA rearrangement)". Development, Growth and Differentiation. 25 (6): 563–569. doi:10.1111/j.1440-169X.1983.00563.x. PMID 37282129. S2CID 83666742.
- ↑ Tsuda T, Yamagishi H, Ohnishi N, Yamada Y, Izumi H, Mori KJ (November 1983). "Extrachromosomal circular DNAs from murine hemopoietic tissue cells". Plasmid. 10 (3): 235–241. doi:10.1016/0147-619X(83)90037-9. PMID 6657775.
- 1 2 Flores SC, Sunnerhagen P, Moore TK, Gaubatz JW (May 1988). "Characterization of repetitive sequence families in mouse heart small polydisperse circular DNAs: age-related studies". Nucleic Acids Research. 16 (9): 3889–3906. doi:10.1093/nar/16.9.3889. PMC 336563. PMID 3375074.
- 1 2 3 4 5 Wang T, Zhang H, Zhou Y, Shi J (June 2021). "Extrachromosomal circular DNA: a new potential role in cancer progression". Journal of Translational Medicine. 19 (1): 257. doi:10.1186/s12967-021-02927-x. PMC 8194206. PMID 34112178.
- 1 2 3 4 5 6 Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, et al. (March 2017). "Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity". Nature. 543 (7643): 122–125. Bibcode:2017Natur.543..122T. doi:10.1038/nature21356. PMC 5334176. PMID 28178237.
- ↑ Møller HD, Mohiyuddin M, Prada-Luengo I, Sailani MR, Halling JF, Plomgaard P, et al. (March 2018). "Circular DNA elements of chromosomal origin are common in healthy human somatic tissue". Nature Communications. 9 (1): 1069. Bibcode:2018NatCo...9.1069M. doi:10.1038/s41467-018-03369-8. PMC 5852086. PMID 29540679.
- 1 2 Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, et al. (November 2019). "Circular ecDNA promotes accessible chromatin and high oncogene expression". Nature. 575 (7784): 699–703. Bibcode:2019Natur.575..699W. doi:10.1038/s41586-019-1763-5. PMC 7094777. PMID 31748743.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 Wang Y, Wang M, Djekidel MN, Chen H, Liu D, Alt FW, Zhang Y (November 2021). "eccDNAs are apoptotic products with high innate immunostimulatory activity". Nature. 599 (7884): 308–314. Bibcode:2021Natur.599..308W. doi:10.1038/s41586-021-04009-w. PMC 9295135. PMID 34671165. S2CID 239051756.
- 1 2 Barreto SC, Uppalapati M, Ray A (May 2014). "Small Circular DNAs in Human Pathology". The Malaysian Journal of Medical Sciences. 21 (3): 4–18. PMC 4163554. PMID 25246831.
- 1 2 3 Dillon LW, Kumar P, Shibata Y, Wang YH, Willcox S, Griffith JD, et al. (June 2015). "Production of Extrachromosomal MicroDNAs Is Linked to Mismatch Repair Pathways and Transcriptional Activity". Cell Reports. 11 (11): 1749–1759. doi:10.1016/j.celrep.2015.05.020. PMC 4481157. PMID 26051933.
- 1 2 Hull RM, King M, Pizza G, Krueger F, Vergara X, Houseley J (December 2019). "Transcription-induced formation of extrachromosomal DNA during yeast ageing". PLOS Biology. 17 (12): e3000471. doi:10.1371/journal.pbio.3000471. PMC 6890164. PMID 31794573.
- 1 2 Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, Regenberg B (October 2010). "Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus". Proceedings of the National Academy of Sciences of the United States of America. 107 (43): 18551–18556. Bibcode:2010PNAS..10718551G. doi:10.1073/pnas.1014023107. PMC 2972935. PMID 20937885.
- 1 2 3 Sinclair DA, Guarente L (December 1997). "Extrachromosomal rDNA circles--a cause of aging in yeast". Cell. 91 (7): 1033–1042. doi:10.1016/s0092-8674(00)80493-6. PMID 9428525. S2CID 12735979.
- ↑ deCarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, et al. (May 2018). "Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma". Nature Genetics. 50 (5): 708–717. doi:10.1038/s41588-018-0105-0. PMC 5934307. PMID 29686388.
- ↑ Kim H, Nguyen NP, Turner K, Wu S, Gujar AD, Luebeck J, et al. (September 2020). "Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers". Nature Genetics. 52 (9): 891–897. doi:10.1038/s41588-020-0678-2. PMC 7484012. PMID 32807987.
- ↑ Kuttler F, Mai S (February 2007). "Formation of non-random extrachromosomal elements during development, differentiation and oncogenesis". Seminars in Cancer Biology. 17 (1): 56–64. doi:10.1016/j.semcancer.2006.10.007. PMID 17116402.
- ↑ Zimmer C (November 20, 2019). "Scientists Are Just Beginning to Understand Mysterious DNA Circles Common in Cancer Cells". New York Times.
- ↑ Jin Y, Liu Z, Cao W, Ma X, Fan Y, Yu Y, et al. (2012). "Novel functional MAR elements of double minute chromosomes in human ovarian cells capable of enhancing gene expression". PLOS ONE. 7 (2): e30419. Bibcode:2012PLoSO...730419J. doi:10.1371/journal.pone.0030419. PMC 3272018. PMID 22319568.
- ↑ Vicario R, Peg V, Morancho B, Zacarias-Fluck M, Zhang J, Martínez-Barriocanal Á, et al. (2015-06-15). "Patterns of HER2 Gene Amplification and Response to Anti-HER2 Therapies". PLOS ONE. 10 (6): e0129876. Bibcode:2015PLoSO..1029876V. doi:10.1371/journal.pone.0129876. PMC 4467984. PMID 26075403.
Further reading
- Sinclair DA, Guarente L (December 1997). "Extrachromosomal rDNA circles--a cause of aging in yeast". Cell. 91 (7): 1033–1042. doi:10.1016/s0092-8674(00)80493-6. PMID 9428525.
- Cohen S, Houben A, Segal D (March 2008). "Extrachromosomal circular DNA derived from tandemly repeated genomic sequences in plants". The Plant Journal. 53 (6): 1027–1034. doi:10.1111/j.1365-313x.2007.03394.x. PMID 18088310.
- Cohen S, Regev A, Lavi S (February 1997). "Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability". Oncogene. 14 (8): 977–985. doi:10.1038/sj.onc.1200917. PMID 9050997.
- Kumar P, Dillon LW, Shibata Y, Jazaeri AA, Jones DR, Dutta A (September 2017). "Normal and Cancerous Tissues Release Extrachromosomal Circular DNA (eccDNA) into the Circulation". Molecular Cancer Research. 15 (9): 1197–1205. doi:10.1158/1541-7786.MCR-17-0095. PMC 5581709. PMID 28550083.
- Baskin F, Rosenberg RN, Dev V (June 1981). "Correlation of double-minute chromosomes with unstable multidrug cross-resistance in uptake mutants of neuroblastoma cells". Proceedings of the National Academy of Sciences of the United States of America. 78 (6): 3654–3658. Bibcode:1981PNAS...78.3654B. doi:10.1073/pnas.78.6.3654. PMC 319629. PMID 6943568. Free full-text.
- Barker PE (February 1982). "Double minutes in human tumor cells". Cancer Genetics and Cytogenetics. 5 (1): 81–94. doi:10.1016/0165-4608(82)90043-7. PMID 6175392.
- Masters J, Keeley B, Gay H, Attardi G (May 1982). "Variable content of double minute chromosomes is not correlated with degree of phenotype instability in methotrexate-resistant human cell lines". Molecular and Cellular Biology. 2 (5): 498–507. doi:10.1128/MCB.2.5.498. PMC 369819. PMID 7110138. Free full-text.