FAM63B is a protein which in humans is encoded by the gene FAM63B. This gene is highly expressed in humans. The FAM63B gene is also highly conserved throughout evolutionary history. The discovered function of FAM63B is an interaction with the kinesin-1 light chain and the transportation of vaccinia virus from the nucleus to the cell periphery.
Gene
Locus
FAM63B is located at 15q21.3-q22.1,[1] spanning 90,707 base pairs on chromosome 15.[2]
![]()
Alternative Names
The full name of FAM63B is family with sequence similarity 63, member B.[3] FAM63B is also listed by its alias, KIAA1164, in some publications.[4]
mRNA
Isoforms
The FAM63B gene encodes a primary transcript that can be alternatively spliced into 9 protein variants. FAM63B variant a is the most common isoform found in humans.[2]
| Variant | Length (amino acids) | Exon Count | Molecular Weight (kdal) | Isoelectric Point |
|---|---|---|---|---|
| a | 621 | 9 | 67.1 | 4.24 |
| b | 620 | 9 | 67.0 | 4.24 |
| x1 | 639 | 10 | 69.2 | 4.40 |
| x2 | 638 | 10 | 69.1 | 4.40 |
| x3 | 605 | 10 | 65.2 | 4.41 |
| x4 | 587 | 9 | 63.1 | 4.25 |
| x5 | 364 | 6 | 38.0 | 4.50 |
| x6 | 351 | 8 | 40.2 | 4.72 |
| x7 | 342 | 8 | 39.1 | 4.45 |
Protein
Structure
Primary Structure
FAM63B is a member of the Pfam super family, and contains a domain of unknown function (DUF544) that is homologous within the protein family.[2] FAM63B protein variant an also contains a bipartite tryptophan binding motif from W476 to W533.[6] Variant a of the protein also contains a hydrophobic stretch of alanine from 567 to 574 and a mixed charge sequence from residue 598 to 617.[5] FAM63B protein may contain a signal sequence specifying return to the endoplasmic reticulum (KDEL) from residue 607 to 621 in variant a.
Secondary Structure
The secondary structure of FAM63B is a combination of coils, some α-helices, and few β-sheets.[5][7][8] The Phyre 2 program predicts α-helices in 23% of the protein, β-strands in 9% of the protein, and the remaining 59% of the protein as disordered.[7] The disordered regions coincide with the coiled regions predicted by other programs, and this results in the long stretch of coiled protein beginning at the N-terminus. According to the SOUSI program, there is a 16-amino acid-long span from residues 265 to 280 of FAM63B that could be a transmembrane sequence.[9] However, transmembrane sequences generally need to be at least 20 amino acids long in order to be stable in the membrane, so a transmembrane sequence is unlikely. Therefore, FAM63B is not fixed in the membrane of any organelle and is free to move through the cell and between organelles.
Tertiary Structure
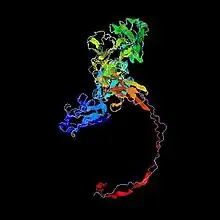
Not much is known about the tertiary structure of FAM63B. A predicted folding is shown.
Post-translational Modifications
Post-translational modifications of the FAM63B protein.[9]
| Post-translational Modification | Site(s) | Impact on Protein |
|---|---|---|
| Acetylation | Ser3 | Stability, localization, metabolism, apoptosis, ribosome recognition for synthesis |
| Lysine Glycation | Lys88, Lys251, Lys280, Lys282, Lys332, Lys393, Lys398, Lys454, Lys547 | Impaired function, changed characteristics |
| Phosphorylation | Ser7, Ser21, Ser25, Ser26, Ser62, Ser66, Ser68, Ser72, Ser90, Ser94, Ser111, Ser148, Ser153, Ser158, Ser160, Ser165, Ser170, Ser175, Ser188, Ser193, Ser233, Ser396, Ser440, Ser499, Ser541, Ser558, Ser587, Ser589, Ser590, Ser594, Ser597, Thr48, Thr255, Thr344, Thr453, Tyr505 | Conformation change, turn enzymatic activity on/off |
| Picornaviral Cleavage | Glu195, Gln535 | Cleavage of polyprotein, degradation |
| O-GlcNAc | Ser3, Ser21, Ser49, Ser62, Ser66, Ser68, Ser80, Ser152, Ser153, Ser158, Ser170, Ser499, Ser575, Ser587, Ser589, Ser590, Thr144, Thr576 | Nucleocytoplasmic location |
Subcellular Location
FAM63B has predicted NES (nuclear export signals) at Val274 and Leu277.[10] Also, a NLS (nuclear localization signal) is predicted for FAM63B at RKRK at residue 599.[9] In agreement, Reinhardt's method for cytoplasmic/nuclear discrimination predicts FAM63B to be located in the nucleus with a reliability of 76.7%. The presence of both NLS and NES signals and O-GlcNAc post-translational modification of FAM63B supports the protein's location in both the nucleus and cytoplasm and the discovered protein function as a shuttle for vaccinia virus between the nucleus and the cell periphery.
Expression
Expression Level
FAM63B has moderately-high to high expression and is constitutively expressed. FAM63B is likely ubiquitously expressed in humans.[11]
Differential Expression
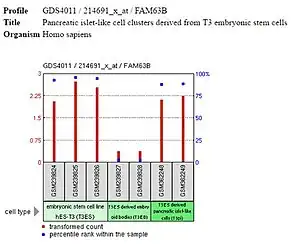
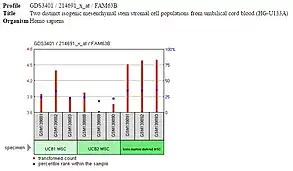
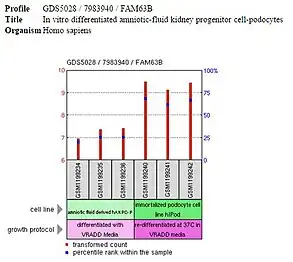
Expression of FAM63B is high in the embryonic stem cells and differentiated tissues but low or off in embryoid bodies and other progenitor cells, such as the multipotent mesenchymal stem cells. It is likely that FAM63B is expressed during pluripotency and unipotency but is not important for differentiation, as is occurring in embryoid bodies, mesenchymal stem cells, and other progenitor cells.
Regulation of Expression
Transcriptional Regulation
The promoter of FAM63B is GXP_5885, located on the positive strand of chromosome 15 from (58770692, 58771462) and is 711 base pairs long.[12]
Interacting Proteins
FAM63B is shown to interact with one protein, KLC-1.[13] KLC-1, kinesin light chain 1, is a protein which recruits kinesin-1 via its cargo binding light chain and contains a bipartite tryptophan binding motif.[13] This motif is present in a vaccinia virus integral membrane protein, A36, that is required for transport of the virus from the perinuclear space to the cell periphery.[13] In the absence of A36, proteins with a bipartite tryptophan binding motif can interact with the kinesin light chain, recruit KLC-1, and promote virus transport from the nucleus to the cytoplasm.[13]
Function
The discovered function of FAM63B protein is a transporter of vaccinia virus in the human genome. FAM63B contains a bipartite tryptophan binding motif between W476 and W533.[13] The motif also contains a Q residue at the +2 position, which was found to be a frequent occurrence in proteins that bind KLC-1 or KLC-2.[13] FAM63B is among proteins studied that can rescue virus transport to the cell periphery when expressed in A36-deficient cells, successfully replacing the cytoplasmic domain A36 of vaccinia.[13]
Clinical Significance
Pathology
The specific pathology of FAM63B is unknown.
Disease Association
FAM63B is part of four networks regulated by miRNA, three of which are linked to neuronal differentiation and dopaminergic gene expression.[14] These findings indicate that FAM63B could be used as a biomarker for the detection and treatment of schizophrenia.[14] Furthermore, aberrant methylation of FAM63B may play a role in the development of schizophrenia.[14] FAM63B has also been ranked 13 of 25 on a list of associated genes relevant to arthritis.[15]
Homology
Paralogs
FAM63B has one paralog, FAM63A, which is a gene of unknown function. FAM63A gene encodes a protein that is 469 amino acids long and 76% similar to FAM63B.[16]
Orthologs
FAM63B has been found in all multicellular and unicellular eukaryotes, including plants but excluding protists and fungi. The gene has also been found in archaea but not bacteria.[17]
| Genus & species | Common Name | Date of Divergence from Humans (MYA) | Accession Number | Sequence Length (amino acids) | Sequence Similarity to Human Protein (%) | Clade |
|---|---|---|---|---|---|---|
| Pan troglodytes | Chimpanzee | 6.2 | XP_510443.2 | 621 | 100 | Mammalia |
| Microtus ochrogaster | Prairie vole | 90.1 | XP_005347720.1 | 597 | 84 | Mammalia |
| Ursus maritimus | Polar bear | 95 | XP_008704293.1 | 591 | 97 | Mammalia |
| Acinonyx jubatus | Cheetah | 95 | XP_014926357.1 | 488 | 96 | Mammalia |
| Pelodiscus sinensis | Green sea turtle | 320.5 | XP_014434471.1 | 408 | 95 | Reptilia |
| Zonotrichia albicollis | White-throated sparrow | 320.5 | XP_014123064.1 | 326 | 91 | Aves |
| Columba livia | Rock dove | 320.5 | XP_005511195.1 | 340 | 91 | Aves |
| Chrysemys picta bellii | Western painted turtle | 320.5 | XP_008162326.1 | 566 | 86 | Reptilia |
| Melopsittacus undulatus | Budgerigar | 320.5 | XP_005145999.1 | 472 | 86 | Aves |
| Xenopus tropicalis | Western clawed frog | 354.4 | XP_002937714.1 | 354 | 87 | Amphibia |
| Latimeria chalumnae | Western Indian Ocean coelacanth | 413.69 | XP_005998789.1 | 652 | 83 | Sarcopterygii |
| Lepisosteus oculatus | Spotted gar | 436.8 | XP_015198676.1 | 642 | 82 | Actinopterygii |
| Hydra vulgaris | Hydra | 902 | XP_012556960.1 | 507 | 65 | Hydrozoa |
| Octopus bimaculoides | California two-spot octopus | 903 | XP_014779548.1 | 981 | 58 | Cephalopoda |
| Crassostrea gigas | Pacific oyster | 903 | XP_011440367.1 | 569 | 71 | Bivalvia |
| Haemonchus contortus | Barber's pole worm | 903 | CDJ97151.1 | 437 | 55 | Secernentea |
| Trichoplax adhaerens | Trichoplax | 936 | XP_002108532.1 | 308 | 75 | Placozoa |
| Solanum pennellii | Tomato | 1570.5 | XP_015085752.1 | 686 | 65 | Angiosperms |
| Sesamum indicum | Sesame | 1570.5 | XP_011071984.1 | 738 | 65 | Angiosperms |
| Thermoplasmatales archaeon | BRNA1 | 4250 | WP_048164282.1 | 1063 | 39 | Archaea |
Distant Homologs
The most distant homolog of FAM63B is found in Thermoplasmatales archaeon, an archaea that diverged from the human gene 4.25 billion years ago.[17][18]
Homologous Domains
FAM63B is a member of the Pfam super family, and contains a domain of unknown function (DUF544) homologous within the protein family.[17] This region of the protein is highly conserved through FAM63B homologs, as is the bipartite tryptophan binding motif of FAM63B and the C-terminus signal sequence.
Phylogeny
The phylogenetic tree below shows a time calibration for the evolution of FAM63B.

References
- ↑ "UCSC Genome Browser".
- 1 2 3 "FAM63B family with sequence similarity 63 member B [Homo sapiens (human)] - Gene - NCBI". nih.gov.
- ↑ "Aliases for FAM63B Gene". GeneCards.
- ↑ "MINDY2 Gene - GeneCards | MINY2 Protein | MINY2 Antibody".
- 1 2 3 http://seqtool.sdsc.edu/CGI/BW.cgi#%5B%5D!
- ↑ Dodding, M. P., Mitter, R., Humphries, A. C., & Way, M. (2011). A kinesin-1 binding motif in vaccinia virus that is widespread throughout the human genome. The EMBO Journal, 30(22), 4523–4538. http://doi.org/10.1038/emboj.2011.326
- 1 2 "Phyre 2 Results for Undefined". ic.ac.uk.
- ↑ "I-TASSER server for protein structure and function prediction". umich.edu.
- 1 2 3 "ExPASy: SIB Bioinformatics Resource Portal - Categories". expasy.org.
- ↑ "5716AACA000019874EC1B1F0 expired". dtu.dk.
- ↑ "Family with sequence similarity 63, member B (FAM63B)". nih.gov.
- ↑ "Genomatix: Annotation & Analysis". genomatix.de.
- 1 2 3 4 5 6 7 Dodding, M. P., Mitter, R., Humphries, A. C., & Way, M. (2011). A kinesin-1 binding motif in vaccinia virus that is widespread throughout the human genome. The EMBO Journal, 30(22), 4523–4538. http://doi.org/10.1038/emboj.2011.326
- 1 2 3 Aberg, K. A., et al. (2014). Methylome-Wide Association Study of Schizophrenia. JAMA Psychiatry, 71(3), 255–264. http://doi.org/10.1001/jamapsychiatry.2013.3730
- ↑ Li, C., et al. (2008). A systematic method for mapping multiple loci: An application to construct a genetic network for rheumatoid arthritis. Gene, 408(1–2), 104–111. http://doi.org/10.1016/j.gene.2007.10.028
- ↑ "FAM63A family with sequence similarity 63 member A [Homo sapiens (human)] - Gene - NCBI". nih.gov.
- 1 2 3 "BLAST: Basic Local Alignment Search Tool". nih.gov.
- ↑ "TimeTree". timetree.org.