| Haplogroup O-M122 | |
|---|---|
| Possible time of origin | 33,943 (95% CI 25,124 <-> 37,631) ybp [1] 35,000 (with slower average mutation rate) or 30,000 (with faster average mutation rate) years ago[2] |
| Coalescence age | 30,365 (95% CI 22,492 <-> 33,956) ybp[1] |
| Possible place of origin | South Asia or Southeast Asia [3] |
| Ancestor | O-M175 |
| Defining mutations | M122 [4] |
| Highest frequencies | Nyishi 94%, [5] Adi 89%,[5] Tamang 87%, [6] Kachari (Boro) 85%,[7] Apatani 82%,[5] Rabha 76.5%,[7] Naga 76%,[5] Bhutanese 74%, Naiman Kazakhs 68%,[8] Han Chinese 56%, Tibetan 48%, She People 48% (78.6% Northern,[9] 62.7%[10]), Manchus 47%, Hmong/Miao 46% (69.0% China,[10] 64.3% Thailand,[11] 44.0% Hunan,[9] 41.2% Laos,[9] 36.7% Yunnan,[9] 30.6% Guizhou,[9] 14.6% Điện Biên Phủ[12]), Vietnamese 44%, Korean 43%, Karen 37%,[13] Filipinos 33%, Southwestern Tai approx. 30.4%[14] (Shan 40%,[15] Siamese 39.5%,[14] Northern Thai 37.2%,[15] Yong 37%,[13] Tai Lue 29%,[15] Saek 29%,[14] Phuan 29%,[14] Thái in Vietnam 29%,[12] Lao 27.5%,[14] Kaleun 24%,[14] Nyaw 22%,[14] Isan 21%,[14] Tai Khün 21%,[15] Phutai 17%,[14] Tai Dam 14%[14]) |
Haplogroup O-M122 (also known as Haplogroup O2 (formerly Haplogroup O3)) is an Eastern Eurasian Y-chromosome haplogroup. The lineage ranges across Southeast Asia and East Asia, where it dominates the paternal lineages with extremely high frequencies. It is also significantly present in Central Asia, especially among the Naiman tribe of Kazakhs.[8]
This lineage is a descendant haplogroup of haplogroup O-M175.
Origins
Researchers believe that O-M122 first appeared in Southeast Asia approximately 25,000-30,000 years ago [3] or roughly between 30,000 and 35,000 years ago according to more recent studies (Karmin et al. 2015, Poznik et al. 2016, YFull January 4, 2018). In a systematic sampling and genetic screening of an East Asian–specific Y-chromosome haplogroup (O-M122) in 2,332 individuals from diverse East Asian populations, results indicate that the O-M122 lineage is dominant in East Asian populations, with an average frequency of 44.3%. Microsatellite data show that the O-M122 haplotypes are more diverse in Southeast Asia than those in northern East Asia.[3] This suggests a southern origin of the O-M122 mutation to be likely.
It was part of the settlement of East Asia. However, the prehistoric peopling of East Asia by modern humans remains controversial with respect to early population migrations and the place of the O-M122 lineage in these migrations is ambivalent.
Distribution
Although Haplogroup O-M122 appears to be primarily associated with ethnic Tibeto-Burman speaking groups inhabiting the Seven Sister States of north eastern India, it also forms a significant component of the Y-chromosome diversity of most modern populations of the East Asian region.
East Asia
Haplogroup O-M122 is found in approximately 53.29% of all modern Chinese males[16] (with frequency ranging from 30/101=29.7% among Pinghua-speaking Hans in Guangxi [17] to 110/148=74.3% among Hans in Changting, Fujian [18]), about 40% of Manchu, Chinese Mongolian, Korean, and Vietnamese males, about 33.3% [19] to 62% (Jin 2009 and [20]) of Filipino males, about 10.5% [21] to 55.6% [21] of Malaysian males, about 10% (4/39 Guide County, Qinghai) [22] to 45% (22/49 Zhongdian County, Yunnan) [23] of Tibetan males, about 20% (10/50 Shuangbai, northern Yunnan) [23] to 44% (8/18 Xishuangbanna, southern Yunnan) [23] and [24] of Yi males, about 25% of Zhuang [25] and Indonesian [26] males, and about 16%[27][28] to 20% [19] of Japanese males. The distribution of Haplogroup O-M122 stretches far into Asia (approx. 40% of Dungans,[29] 30% of Salars,[30] 28% of Bonan,[30] 24% of Dongxiang,[30] 18% to 22.8% of Mongolian citizens in Ulaanbaatar,[19] 11%-15.4% of Khalkha Mongolians (Yamamoto et al. 2013[31]) but also as high as 31.1% (Kim et al. 2011), 12% of Uyghurs,[29] 9% of Kazakhs [29] but in the Naiman of Kazakhs 65.81%,[8] 6.8% of Kalmyks[32] (17.1% of Khoshuud, 6.1% of Dörwöd, 3.3% of Torguud, 0% of Buzawa), 6.2% of Altaians,[33] 5.3% of Kyrgyz,[34] 4.1% of Uzbeks,[29] and 4.0% of Buryats.[35]
Modern northern Han Chinese Y haplogroups and mtdna match those of ancient northern Han Chinese ancestors 3,000 years ago from the Hengbei archeological site. 89 ancient samples were taken. Y haplogroups O3a, O3a3, M, O2a, Q1a1, and O* were all found in Hengbei samples.[36] Three men who lived in the Neolithic era are the ancestors of 40% of Han Chinese, with their Y haplogroups being subclades of O3a-M324 and they are estimated to have lived 6,800 years ago, 6,500 years ago and 5,400 years ago.[37]
The East Asian O3-M122 Y chromosome Haplogroup is found in large quantities in other Muslims close to the Hui people like Dongxiang, Bo'an and Salar. The majority of Tibeto-Burmans, Han Chinese, and Ningxia and Liaoning Hui share paternal Y chromosomes of East Asian origin which are unrelated to Middle Easterners and Europeans. In contrast to distant Middle Eastern and Europeans whom the Muslims of China are not related to, East Asians, Han Chinese, and most of the Hui and Dongxiang of Linxia share more genes with each other. This indicates that native East Asian populations converted to Islam and were culturally assimilated to these ethnicities and that Chinese Muslim populations are mostly not descendants of foreigners as claimed by some accounts while only a small minority of them are.[38]
South Asia
Haplogroup O-M122 is restricted among tribal groups of Northeast India where it is found at very high frequencies. In Arunachal Pradesh, it is found at 89% among Adi, 82% among Apatani, and 94% among Nishi, while the Naga people show it at 100% (Cordaux 2004). In Meghalaya, 59.2% (42/71) of a sample of Garos and 31.7% (112/353) of a sample of Khasis have been found to belong to O-M122.[39] In Nepal, Tamang people present a very high frequency of O-M122 (39/45 = 86.7%), while much lower percentages of Newar (14/66 = 21.2%) and the general population of Kathmandu (16/77 = 20.8%) belong to this haplogroup.[40] A study published in 2009 found O-M122 in 52.6% (30/57, including 28 members of O-M117 and two members of O-M134(xM117)) of a sample of Tharus from a village in Chitwan District of south-central Nepal, 28.6% (22/77, all O-M117) of a sample of Tharus from another village in Chitwan District, and 18.9% (7/37, all O-M117) of a sample of Tharus from a village in Morang District of southeastern Nepal.[41] In contrast, the same study found O-M122 in only one individual in a sample of non-Tharu Hindus collected in Chitwan District (1/26 = 3.8% O-M134(xM117)), one tribal individual from Andhra Pradesh, India (1/29 = 3.4% O-M117), and one individual in a sample of Hindus from New Delhi, India (1/49 = 2.0% O-M122(xM134)).[41]
Southeast Asia
Among all the populations of East and Southeast Asia, Haplogroup O-M122 is most closely associated with those that speak a Sinitic, Tibeto-Burman, or Hmong–Mien language. Haplogroup O-M122 comprises about 50% or more of the total Y-chromosome variation among the populations of each of these language families. The Sinitic and Tibeto-Burman language families are generally believed to be derived from a common Sino-Tibetan protolanguage, and most linguists place the homeland of the Sino-Tibetan language family somewhere in northern China. The Hmong–Mien languages and cultures, for various archaeological and ethnohistorical reasons, are also generally believed to have derived from a source somewhere north of their current distribution, perhaps in northern or central China. The Tibetans, however, despite the fact that they speak a language of the Tibeto-Burman language family, have high percentages of the otherwise rare haplogroups D-M15 and D3, which are also found at much lower frequencies among the members of some other ethnic groups in East Asia and Central Asia.
Haplogroup O-M122 has been implicated as a diagnostic genetic marker[42] of the Austronesian expansion when it is found in populations of insular Southeast Asia and Oceania. It appears at moderately high frequencies in the Philippines, Malaysia, and Indonesia. Its distribution in Oceania is mostly limited to the traditionally Austronesian culture zones, chiefly Polynesia (approx. 25% [19] to 32.5% [21]). O-M122 is found at generally lower frequencies in coastal and island Melanesia, Micronesia, and Taiwanese aboriginal tribes (18% [19] to 27.4% [21] of Micronesians, and 5% of Melanesians,[43] albeit with reduced frequencies of most subclades.
Haplogroup O-M122* Y-chromosomes, which are not defined by any identified downstream markers, are actually more common among certain non-Han Chinese populations than among Han Chinese ones, and the presence of these O-M122* Y-chromosomes among various populations of Central Asia, East Asia, and Oceania is more likely to reflect a very ancient shared ancestry of these populations rather than the result of any historical events. It remains to be seen whether Haplogroup O-M122* Y-chromosomes can be parsed into distinct subclades that display significant geographical or ethnic correlations.
Subclade Distribution
Paragroup O-M122*
Paragroup O2*-M122(xO2a-P197) Y-DNA is quite rare, having been detected only in 2/165 = 1.2% of a sample of Han Chinese in a pool of samples from mainland China, Taiwan, the Philippines, Vietnam, and Malaysia (n=581), 8/641 = 1.2% of a sample of Balinese in a pool of samples from western Indonesia (n=960), and 7/350 = 2.0% of a sample of males from Sumba in a pool of samples from eastern Indonesia (n=957). In the same study, O2*-M122(xO2a-P197) Y-DNA was not observed in a pool of samples from Oceania (n=182).[44]
A paper published by a group of mainly Chinese geneticists in the American Journal of Human Genetics in 2005 reported the detection of O2*-M122(xO2a-M324) Y-DNA in 1.6% (8/488) of a pool of seven samples of Han Chinese (3/64 = 4.7% Sichuan, 2/98 = 2.0% Zibo, Shandong, 1/60 = 1.7% Inner Mongolia, 1/81 = 1.2% Yunnan, 1/86 = 1.2% Laizhou, Shandong, 0/39 Guangxi, 0/60 Gansu). O2*-M122(xO2a-M324) Y-DNA also was detected in the following samples of ethnic minorities in China: 5.9% (1/17) Jingpo from Yunnan, 4.3% (2/47) Zhuang from Yunnan, 4.1% (2/49) Lisu from Yunnan, 3.2% (1/31) Wa from Yunnan, 2.6% (1/39) Zhuang from Guangxi, 2.5% (2/80) Bai from Yunnan, 2.4% (1/41) Hani from Yunnan, 2.3% (2/88) Lahu from Yunnan, 2.1% (1/47) Yi from Yunnan, 2.1% (1/48) Miao from Yunnan, 1.5% (2/132) Dai from Yunnan, 1.0% (1/105) Miao from Hunan, and 0.9% (2/225) Yao from Guangxi.[45]
O2*-M122(xO2a-M324) Y-DNA has been found as a singleton (1/156 = 0.6%) in a sample from Tibet.[40] It also has been found as a singleton in a sample of nineteen members of the Chin people in Chin State, Myanmar.[46]
In a paper published in 2011, Korean researchers have reported finding O2*-M122(xO2a-M324) Y-DNA in the following samples: 5.9% (3/51) Beijing Han, 3.1% (2/64) Filipino, 2.1% (1/48) Vietnamese, 1.7% (1/60) Yunnan Han, 0.4% (2/506) Korean, including 1/87 from Jeju and 1/110 from Seoul-Gyeonggi.[47] In another study published in 2012, Korean researchers have found O-M122(xM324) Y-DNA in 0.35% (2/573) of a sample from Seoul; however, no individual belonging to O-M122(xM324) was observed in a sample of 133 individuals from Daejeon.[48]
In 2011, Chinese researchers published a paper reporting their finding of O2*-M122(xO2a-M324) Y-DNA in 3.0% (5/167) of a sample of Han Chinese with origins in East China (defined as consisting of Jiangsu, Zhejiang, Shanghai, and Anhui) and in 1.5% (1/65) of a sample of Han Chinese with origins in Southern China. O2* Y-DNA was not detected in their sample of Han Chinese with origins in Northern China (n=129).
In a paper published in 2012, O2*-M122(xO2a-P200) Y-DNA was found in 12% (3/25) of a sample of Lao males from Luang Prabang, Laos. O2* Y-DNA was not detected in this study's samples of Cham from Binh Thuan, Vietnam (n=59), Kinh from Hanoi, Vietnam (n=76), or Thai from northern Thailand (n=17).[49]
Trejaut et al. (2014) found O2-M122(xO2a-M324) in 6/40 (15.0%) Siraya in Kaohsiung, 1/17 (5.9%) Sulawesi, 1/25 (4.0%) Paiwan, 2/55 (3.6%) Fujian Han, 1/30 (3.3%) Ketagalan, 2/60 (3.3%) Taiwan Minnan, 1/34 (2.9%) Taiwan Hakka, 1/38 (2.6%) Siraya in Hwalien, 5/258 (1.9%) miscellaneous Han volunteers in Taiwan, and 1/75 (1.3%) in a sample of the general population of Thailand.[50]
Brunelli et al. (2017) found O2-M122(xO2a-M324) in 5/66 (7.6%) Tai Yuan, 1/91 (1.1%) Tai Lue, and 1/205 (0.5%) Khon Mueang in samples of the people of Northern Thailand.[15]
O-M324
O-M121
O2a1a1a1a1-M121 is a subclade of O2a1-L127.1, parallel to O2a1b-M164 and O2a1c-JST002611.
In an early survey of Y-DNA variation in present-day human populations of the world, O-M121 was detected only in 5.6% (1/18) of a sample from Cambodia and Laos and in 5.0% (1/20) of a sample from China.[51]
In a large study of 2,332 unrelated male samples collected from 40 populations in East Asia (and especially Southwest China), O-M121/DYS257 Y-DNA was detected only in 7.1% (1/14) of a sample of Cambodians and in 1.0% (1/98) of a sample of Han Chinese from Zibo, Shandong.[45]
In a study published in 2011, O-M121 Y-DNA was found in 1.2% (2/167) of a sample of Han Chinese with origins in East China, defined as consisting of Jiangsu, Anhui, Zhejiang, and Shanghai, and in 0.8% (1/129) of a sample of Han Chinese with origins in Northern China. O-M121 was not detected in this study's sample of Han Chinese with origins in Southern China (n=65).[52]
O-L599 (considered to be phylogenetically equivalent to O-M121[53]) also has been found in one individual in the 1000 Genomes Project sample of Han Chinese from Hunan, China (n=37), one individual in the 1000 Genomes Project sample of Kinh from Ho Chi Minh City, Vietnam, one individual in the Human Genome Diversity Project sample of Tujia, an individual from Singapore, and an individual from the Jakarta metropolitan area.[54] According to 23mofang, O-L599 currently accounts for about 0.79% of the male population in China and is concentrated in Fujian, Taiwan, Jiangxi, Anhui, Hubei, Zhejiang and other provinces and cities; it appears to have undergone explosive population growth between about 2600 and 2300 years ago.[55]
O-M164
O2a1b-M164 is a subclade of O2a1-L127.1, parallel to O2a1a1a1a1-M121 and O2a1c-JST002611.
In an early survey of Y-DNA variation in present-day human populations of the world, O-M164 was detected only in 5.6% (1/18) of a sample from Cambodia and Laos.[51]
In a large study of 2,332 unrelated male samples collected from 40 populations in East Asia (and especially Southwest China), O2a1b-M164 Y-DNA was detected only in 7.1% (1/14) of a sample of Cambodians.[45]
According to 23mofang, O-M164 is a recent branch (TMRCA 2120 years) downstream of O2a1c-JST002611 rather than parallel to it. Out of fourteen members total, six are from Guangdong, five are from Fujian, one is from Nantong, one is from Wenzhou, and one is from Taiwan.[55]
O-JST002611
Haplogroup O2a1c-JST002611 is derived from O2-M122 via O2a-M324/P93/P197/P199/P200 and O2a1-L127.1/L465/L467. O2a1c-JST002611 is the most commonly observed type of O2a1 Y-DNA, and, more generally, represents the majority of extant O2-M122 Y-DNA that does not belong to the expansive subclade O2a2-P201.
Haplogroup O2a1c-JST002611 was first identified in 3.8% (10/263) of a sample of Japanese (Nonaka et al. 2007). It also has been found in 3.5% (2/57) of the JPT (Japanese in Tokyo, Japan) sample of the 1000 Genomes Project, including one member of the rare and deeply divergent paragroup O2a1c1-F18*(xO2a1c1a1-F117, O2a1c1a2-F449).[54][2] Subsequently, this haplogroup has been found with higher frequency in some samples taken in and around China, including 12/58 = 20.7% Miao (China), 10/70 = 14.3% Vietnam, 18/165 = 10.9% Han (China & Taiwan), 4/49 = 8.2% Tujia (China).[44] O-002611 also has been found in a singleton from the Philippines (1/48 = 2.1%), but it has not been detected in samples from Malaysia (0/32), Taiwanese Aboriginals (0/48), She from China (0/51), Yao from China (0/60), Oceania (0/182), eastern Indonesia (0/957), or western Indonesia (0/960).[44] Haplogroup O2a1c‐JST002611 is prevalent in different ethnic groups in China and Southeast Asia, including Vietnam (14.29%), Sichuan of southwestern China (Han, 14.60%; Tibetan in Xinlong County, 15.22%),[56] Jilin of northeastern China (Korean, 9.36%), Inner Mongolia (Mongolian, 6.58%), and Gansu of northwestern China (Baima, 7.35%; Han, 11.30%).[57] Y-DNA belonging to haplogroup O-JST002611 has been observed in 10.6% (61/573) of a sample collected in Seoul and 8.3% (11/133) of a sample collected in Daejeon, South Korea.[48][58]
According to 23mofang, haplogroup O-IMS-JST002611 currently accounts for approximately 14.72% of the entire male population of China, and its TMRCA is estimated to be 13,590 years.[59] Yan et al. (2011) have found O-IMS-JST002611 in 16.9% (61/361) of a pool of samples of Han Chinese from East China (n=167), North China (n=129), and South China (n=65).[60] According to Table S4 of He Guanglin et al. 2023, haplogroup O2a1b-IMS-JST002611 has been found in 17.50% (366/2091) of a pool of samples of Han Chinese from various provinces and cities of China.[61] Haplogroup O2a1b-IMS-JST002611 is the second most common Y-DNA haplogroup among Han Chinese (and among Chinese in general) after haplogroup O2a2b1a1-M117.
O-P201
O2a2-JST021354/P201 has been divided into primary subclades O2a2a-M188 (TMRCA 18,830 ybp, accounts for approximately 4.74% of all males in present-day China[62]) and O2a2b-P164 (TMRCA 20,410 ybp, accounts for approximately 30.4% of all males in present-day China[63][64]). Among the various branches of O2a2a-M188, O-M7 (TMRCA 14,510 ybp, accounts for approximately 2.15% of all males in present-day China[65]) is notable for its relatively high frequency over a wide swath of Southeast Asia and southern China, especially among certain populations that currently speak Hmong-Mien, Austroasiatic, or Austronesian languages. Other branches of O2a2a-M188, such as O-CTS201 (TMRCA 16,070 ybp, accounts for approximately 1.76% of all males in present-day China[66]), O-MF39662 i.e. O-F2588(xCTS445), and O-MF109044 i.e. O-M188(xF2588) (TMRCA 9,690 ybp, accounts for approximately 0.4% of all males in present-day China[67]) have been found with generally low frequency in China; however, the O-CTS201 > O-FGC50590 > O-MF114497 subclade is fairly common among males in Korea and Japan. O2a2b-P164 has been divided cleanly into O2a2b1-M134 (TMRCA 17,450 ybp, accounts for approximately 27.61% of all males in present-day China[68]), which has been found with high frequency throughout East Asia and especially among speakers of Sino-Tibetan languages, and O2a2b2-AM01822 (TMRCA 16,000 ybp, accounts for approximately 2.79% of all males in present-day China[69]), which has been found with relatively low frequency but high diversity throughout East Asia and with high frequency in Austronesia.
O2a2-P201(xO2a2a1a2-M7, O2a2b1-M134) Y-DNA has been detected with high frequency in many samples of Austronesian-speaking populations, in particular some samples of Batak Toba from Sumatra (21/38 = 55.3%), Tongans (5/12 = 41.7%), and Filipinos (12/48 = 25.0%). [44] Outside of Austronesia, O2a2-P201(xO2a2a1a2-M7, O2a2b1-M134) Y-DNA has been observed in samples of Tujia (7/49 = 14.3%), Han Chinese (14/165 = 8.5%), Japanese (11/263 = 4.2%), Miao (1/58 = 1.7%), and Vietnam (1/70 = 1.4%) (Karafet 2010 and Nonaka 2007).
O-M159
O2a2a1a1a-M159 is a subclade of O2a2-P201 and O2a2a1a1-CTS201. In an early survey of Y-DNA variation in present-day human populations of the world, O-M159 was detected only in 5.0% (1/20) of a sample from China.[51]
Unlike its phylogenetic siblings, O-M7 and O-M134, O-M159 is very rare, having been found only in 2.9% (1/35) of a sample of Han males from Meixian, Guangdong in a study of 988 males from East Asia.[70]
In a study published in 2011, O-M159 was detected in 1.5% (1/65) of a sample of Han Chinese with origins in Southern China. O-M159 was not detected in the same study's samples of Han Chinese with origins in East China (n=167) or Northern China (n=129).[52]
Trejaut et al. (2014) found O-M159 in 5.0% (3/60) Minnan in Taiwan, 4.2% (1/24) Hanoi, Vietnam, 3.88% (10/258) miscellaneous Han volunteers in Taiwan, 3.6% (2/55) Han in Fujian, 3.24% (12/370) Plains Aborigines in Taiwan (mostly assimilated to Han Chinese), 1.04% (2/192) Western Indonesia (1/25 Kalimantan, 1/26 Sumatra), and 0.68% (1/146) Philippines (1/55 South Luzon).[50]
Kutanan et al. (2019) found O-M159 in 1.6% (2/129) of their samples of Thai people from Central Thailand.[14]
According to 23mofang, the TMRCA of haplogroup O-M159 is estimated to be 8,900 years. It is currently distributed mainly in southern China, accounting for about 0.80% of the total male population of China.[71]
O-M7
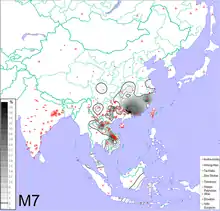
Haplogroup O2a2a1a2-M7 Y-DNA has been detected with high frequency in some samples of populations who speak Hmong-Mien languages, Katuic languages, or Bahnaric languages, scattered through some mostly mountainous areas of southern China, Laos, and Vietnam.[73]
O-M7 has been noted for having a widespread but uneven distribution among populations that speak Hmong-Mien languages, such as She (29/51 = 56.9% She, 10/34 = 29.4% She, 14/56 = 25.0% Northern She from Zhejiang), Miao (21/58 = 36.2% Miao from China, 17/51 = 33.3% Hmong Daw from northern Laos, 6/49 = 12.2% Yunnan Miao, 2/49 = 4.1% Guizhou Miao, 4/100 = 4.0% Hunan Miao), and Yao (18/35 = 51.4% Yao from Liannan, Guangdong, 29/60 = 48.3% Yao from Guangxi, 12/35 = 34.3% Yao from Bama, Guangxi, 12/37 = 32.4% Zaomin from Guangdong, 5/36 = 13.9% Bunu from Guangxi, 1/11 = 9.1% Top-Board Mien, 3/41 = 7.3% Native Mien, 2/31 = 6.5% Southern Mien from Guangxi, 1/19 = 5.3% Flowery-Headed Mien from Guangxi, 1/20 = 5.0% Mountain Straggler Mien from Hunan, 1/28 = 3.6% Blue Kimmun from Guangxi, 1/31 = 3.2% Pahng from Guangxi, 1/47 = 2.1% Western Mien from Yunnan, 0/11 Thin Board Mien, 0/31 Lowland Yao from Guangxi, 0/32 Mountain Kimmun from Yunnan, 0/33 Northern Mien, and 0/41 Lowland Kimmun from Guangxi). [73][44][70]
Cai et al. 2010 have reported finding high frequencies of O-M7 in their samples of Katuic (17/35 = 48.6% Ngeq, 10/45 = 22.2% Katu, 6/37 = 16.2% Kataang, 3/34 = 8.8% Inh (Ir), 4/50 = 8.0% So, 1/39 = 2.6% Suy) and Bahnaric (15/32 = 46.9% Jeh, 17/50 = 34.0% Oy, 8/32 = 25.0% Brau, 8/35 = 22.9% Talieng, 4/30 = 13.3% Alak, 6/50 = 12.0% Laven) peoples from southern Laos. However, O-M7 has been found only with low frequency in samples of linguistically related Khmuic populations from northern Laos (1/50 = 2.0% Mal,[9] 1/51 = 2.0% Khmu,[9] 0/28 Bit,[9] 0/29 Xinhmul[9]), Vietic peoples from Vietnam and central Laos (8/76 = 10.5% Kinh from Hanoi, Vietnam,[74] 4/50 = 8.0% Kinh from northern Vietnam,[12] 2/28 = 7.1% Bo,[9] 4/70 = 5.7% Vietnamese,[10] 0/12 Muong,[9] 0/15 Kinh,[9] 0/38 Aheu[9]), Palaungic peoples from northwestern Laos and southwestern Yunnan (2/35 = 5.7% Lamet,[9] 0/29 Ava,[9] 0/52 Blang[9]), and Pakanic peoples from southeastern Yunnan and northwestern Guangxi (0/30 Palyu,[9] 0/32 Bugan[9]).[73][44][49]
Haplogroup O-M7 has been found with notable frequency in some samples of Austronesian populations from the central part of the Malay Archipelago (17/86 = 19.8% Indonesians from Borneo,[10] 4/32 = 12.5% Malaysia,[10] 7/61 = 11.5% Java (mostly sampled in Dieng),[10] 6/56 = 10.7% Sumatra,[75] 4/53 = 7.5% Java,[75] 1/17 = 5.9% Malaysia[75]), but the frequency of this haplogroup appears to drop off very quickly toward the east (1/48 = 2.1% Philippines,[10] 5/641 = 0.8% Balinese,[10] 0/9 Timor,[10] 0/28 Alor,[10] 0/30 Moluccas,[10] 0/31 Nusa Tenggaras,[75] 0/33 Moluccas,[75] 0/37 Philippines,[75] 0/40 Borneo,[75] 0/48 Taiwanese Aboriginals,[10] 0/54 Mandar from Sulawesi,[10] 0/92 Lembata,[10] 0/350 Sumba,[10] 0/394 Flores[10]) and toward the west (0/38 Batak Toba from Sumatra,[10] 0/60 Nias,[10] 0/74 Mentawai[10]). O-M7 has been found in 14.8% (4/27) of a sample of Giarai from southern Vietnam,[12] 8.3% (2/24) of a sample of Ede from southern Vietnam,[12] and 5.1% (3/59) of a sample of Cham from Binh Thuan, Vietnam.[74] These Chamic-speaking peoples inhabit southern Vietnam and eastern Cambodia, but their languages are related to those of the Acehnese and Malays. O-M7 also has been found in 21.1% (8/38) of a small set of samples of highlanders of northern Luzon (including 1/1 Ifugao, 1/2 Ibaloi, 4/12 Kalangoya, and 2/6 Kankanaey).[76]
In the northern fringes of its distribution, O-M7 has been found in samples of Oroqen (2/31 = 6.5%), Tujia from Hunan (3/49 = 6.1%), Qiang (2/33 = 6.1%), Han Chinese (2/32 = 6.3% Han from Yili, Xinjiang, 4/66 = 6.1% Han from Huize, Yunnan, 2/35 = 5.7% Han from Meixian, Guangdong, 1/18 = 5.6% Han from Wuhan, Hubei, 6/148 = 4.1% Han from Changting, Fujian, 20/530 = 3.8% Han Chinese from Chongming Island,[77] 2/63 = 3.2% Han from Weicheng, Sichuan, 18/689 = 2.6% Han Chinese from Pudong,[77] 2/100 = 2.0% Han from Nanjing, Jiangsu, 3/165 = 1.8% Han Chinese,[44] 1/55 = 1.8% Han from Shanghai),[23][70] Manchus (1/50 = 2.0% Manchu from Liaoning[78]), and Koreans (2/133 = 1.5% Daejeon,[48] 1/300 = 0.3% unrelated Korean males obtained from the National Biobank of Korea,[79] 1/573 = 0.2% Seoul[48]).
According to 23mofang, O-M7 has a TMRCA of approximately 14,510 years and is currently relatively common among many ethnic groups in Sichuan and Yunnan, as well as among the Zhuang, Austroasiatic, and Austronesian groups. O-M7 now accounts for about 2.15% of the total male population in China.[55] The O-N5 subclade (TMRCA 4,230 ybp) by itself accounts for about 0.40% of the total male population in China at present, with its proportion among Hmong-Mien-speaking populations in Southwest China being rather high; in regard to geography, it is found mainly in Guizhou (3.52% of the total provincial population), Hunan (1.63%), Chongqing (1.05%), Sichuan (0.83%), Guangxi (0.76%), Fujian (0.44%), Yunnan (0.35%), Guangdong (0.28%), Jiangxi (0.26%), Hubei (0.26%), Shaanxi (0.20%), and Ningxia (0.18%).[80]
O-M134
O-M134*
Paragroup O-M134(xM117) has been found with very high frequency in some samples of Kim Mun people, a subgroup of the Yao people of southern China (16/32 = 50.0% Mountain Kimmun from southern Yunnan, 11/28 = 39.3% Blue Kimmun from western Guangxi). However, this paragroup has been detected in only 3/41 = 7.3% of a sample of Lowland Kimmun from eastern Guangxi.[73] This paragroup also has been found with high frequency in some Kazakh samples, especially the Naiman tribe (102/155 = 65.81%)(Dulik 2011) Dulik hypothesizes that O-M134 in Kazakhs was due to a later expansion due to its much more recent TMRCA time.
The general outline of the distribution of O-M134(xM117) among modern populations is different as that of the related clade O-M117. In particular, O-M134(xM117) occurs with only low frequency or is nonexistent among most Tibeto-Burman-speaking populations of Southwest China, Northeast India, and Nepal, who exhibit extremely high frequencies of O-M117. This paragroup also occurs with very low frequency or is non-existent among most Mon-Khmer population of Laos, who exhibit much higher frequencies of O-M117.[73] In Han Chinese, the paragroup is found in approximately the same percentage as O-M117, but has a higher distribution in northern Han Chinese than Southern Han Chinese.
According to 23mofang, the TMRCA of O-M134 is estimated to be 17,450 years, and O-M134(xM117) can be divided into two subsets: O-F122 (TMRCA 17,420 years), which is subsumed alongside O-M117 in an O-F450 clade (TMRCA 17,430 years), and O-MF59333 (TMRCA 13,900 years, currently distributed mainly in southern China and accounting for the Y-DNA of approximately 0.03% of the total male population of China), which is derived from O-M134 but basal to O-F450. O-F122 in turn is divided into O-MF38 (TMRCA 4,680 years, currently distributed mainly in northern China and accounting for the Y-DNA of approximately 0.02% of the total male population of China) and O-F114 (TMRCA 15,320 years, accounts for the Y-DNA of approximately 11.29% of the total male population of China).[55] The O-F46 (TMRCA 10,050 years) subclade of O-F114 by itself accounts for the Y-DNA of approximately 10.07% of the total male population of present-day China.[55]
In a study of Koreans from Seoul (n=573) and Daejeon (n=133), haplogroup O-M134(xM117), all members of which have been found to belong to O-F444[58] (phylogenetically equivalent to O-F114[55]), has been found in 9.42% of the sample from Seoul and 10.53% of the sample from Daejeon.[48]
In a study of Japanese (n=263), haplogroup O-M134(xM117) has been observed in nine individuals, or 3.4% of the entire sample set.[81] The Japanese members of O-M134(xM117) in this study have originated from Shizuoka (3/12 = 25%), Tokyo (2/52 = 3.8%), Toyama (1/3), Ishikawa (1/4), Tochigi (1/5), and Ibaraki (1/5), respectively.[82]
O-M117
Haplogroup O2a2b1a1-M117 (also defined by the phylogenetically equivalent mutation Page23) is a subclade of O2a2b1-M134 that occurs frequently in China and in neighboring countries, especially among Tibeto-Burman-speaking peoples. Haplogroup O2a2b1a1-M117 is the most common Y-DNA haplogroup among present-day Chinese (16.27% China,[83] 59/361 = 16.3% Han Chinese,[60] 397/2091 = 18.99% Han Chinese[61]), followed closely by haplogroup O2a1b-IMS-JST002611.
O-M117 has been detected in samples of Tamang (38/45 = 84.4%),Tibetans (45/156 = 28.8% or 13/35 = 37.1%), Tharus (57/171 = 33.3%), Han Taiwanese (40/183 = 21.9%), Newars (14/66 = 21.2%), the general population of Kathmandu, Nepal (13/77 = 16.9%), Han Chinese (5/34 = 14.7% Chengdu, 5/35 = 14.3% Harbin, 4/35 = 11.4% Meixian, 3/30 = 10.0% Lanzhou, 2/32 = 6.3% Yili), Tungusic peoples from the PRC (7/45 = 15.6% Hezhe, 4/26 = 15.4% Ewenki, 5/35 = 14.3% Manchu, 2/41 = 4.9% Xibe, 1/31 = 3.2% Oroqen), Koreans (4/25 = 16.0% Koreans from the PRC, 5/43 = 11.6% Koreans from South Korea), Mongols (5/45 = 11.1% Inner Mongolian, 3/39 = 7.7% Daur, 3/65 = 4.6% Outer Mongolian), and Uyghurs (2/39 = 5.1% Yili, 1/31 = 3.2% Urumqi) (Xue 2006, Gayden 2007, and Fornarino 2009).
Like O-M7, O-M117 has been found with greatly varying frequency in many samples of Hmong-Mien-speaking peoples, such as Mienic peoples (7/20 = 35.0% Mountain Straggler Mien, 9/28 = 32.1% Blue Kimmun, 6/19 = 31.6% Flower Head Mien, 3/11 = 27.3% Top Board Mien, 3/11 = 27.3% Thin Board Mien, 11/47 = 23.4% Western Mien, 6/33 = 18.2% Northern Mien, 5/31 = 16.1% Lowland Yao, 5/35 = 14.3% Yao from Liannan, Guangdong, 5/37 = 13.5% Zaomin, 5/41 = 12.2% Lowland Kimmun, 3/41 = 7.3% Native Mien, 2/31 = 6.5% Southern Mien, 2/32 = 6.3% Mountain Kimmun, but 0/35 Yao from Bama, Guangxi), She (6/34 = 17.6% She, 4/56 = 7.1% Northern She), and Hmongic peoples (9/100 = 9.0% Miao from Hunan, 4/51 = 7.8% Hmong Daw from northern Laos, 3/49 = 6.1% Miao from Yunnan, 1/49 = 2.0% Miao from Guizhou, but 0/36 Bunu from Guangxi) (Cai 2011 and Xue 2006).
In a study published by Chinese researchers in the year 2006, O-M117 has been found with high frequency (8/47 = 17.0%) in a sample of Japanese that should be from Kagawa Prefecture according to the geographical coordinates (134.0°E, 34.2°N) that have been provided.[70] However, in a study published by Japanese researchers in the year 2007, the same haplogroup has been found with much lower frequency (11/263 = 4.2%) in a larger sample of Japanese from various regions of Japan.[28] More precisely, the Japanese members of O-M117 in this study's sample set have originated from Tokyo (4/52), Chiba (2/44), Gifu (1/2), Yamanashi (1/2), Hiroshima (1/3), Aichi (1/6), and Shizuoka (1/12).[82]
In Meghalaya, a predominantly tribal state of Northeast India, O-M133 has been found in 19.7% (14/71) of a sample of the Tibeto-Burman-speaking Garos, but in only 6.2% (22/353, ranging from 0/32 Bhoi to 6/44 = 13.6% Pnar) of a pool of eight samples of the neighboring Khasian-speaking tribes.[39]
O-M300
O-M333
| Population | Frequency | n | Source | SNPs |
|---|---|---|---|---|
| Derung | 1 | Shi 2009 | ||
| Naga (Sagaing, Myanmar) | 1.000 | 15 | Page23=15 | |
| Nishi | 0.94 | Cordaux 2004 | ||
| Adi | 0.89 | Cordaux 2004 | ||
| Tamang | 0.867 | 45 | Gayden 2007 | M134 |
| Nu | 0.86 | Wen 2004 | ||
| Yao (Liannan) | 0.829 | 35 | Xue 2006 | M7=18 M117=5 M122(xM159, M7, M134)=4 M134(xM117)=2 |
| Achang | 0.825 | Shi 2009 | ||
| Apatani | 0.82 | Cordaux 2004 | ||
| Bai | 0.82 | Shi 2009 | ||
| CHS (Han in Hunan & Fujian) | 0.788 | 52 | Poznik 2016 | M122=41 |
| Naga (NE India) | 0.765 | 34 | Cordaux 2004 | M134=26 |
| Ava (Yunnan) | 0.759 | 29 | Cai 2011 | M122 |
| Han Chinese | 0.74 | Wen 2004 | ||
| She | 0.735 | 34 | Xue 2006 | M7=10 M122(xM159, M7, M134)=7 M117=6 M134(xM117)=2 |
| Nu | 0.7 | Shi 2009 | ||
| Miao | 0.7 | Karafet 2001 | ||
| Shui | 0.7 | Shi 2009 | ||
| Han (Harbin) | 0.657 | 35 | Xue 2006 | M122(xM159, M7, M134)=10 M134(xM117)=8 M117=5 |
| Lisu | 0.65 | Wen 2004 | ||
| Zaomin (Guangdong) | 0.649 | 37 | Cai 2011 | M122 |
| She | 0.63 | Karafet 2001 | ||
| Filipinos | 0.62 | Jin 2009 | ||
| Taiwan Han | 0.619 | 21 | Tajima 2004 | M122 |
| Philippines | 0.607 | 28 | Hurles 2005 | M122 |
| Han (East China) | 0.593 | 167 | Yan 2011 | M122 |
| Garo | 0.59 | Reddy 2007 | ||
| Kinh (Hanoi, Vietnam) | 0.58 | 48 | M122=28 | |
| Chin (Chin State, Myanmar) | 0.579 | 19 | Page23=10 M122(xM324)=1 | |
| Han (North China) | 0.566 | 129 | Yan 2011 | M122 |
| Toba (Sumatra) | 0.553 | 38 | Karafet 2010 | P201(xM7, M134) |
| Northern Han | 0.551 | 49 | Tajima 2004 | M122 |
| Garo | 0.55 | Kumar 2007 | ||
| Tujia | 0.54 | Shi 2009 | ||
| Tujia | 0.53 | Karafet 2001 | ||
| Han (Chengdu) | 0.529 | 34 | Xue 2006 | M122(xM159, M7, M134)=8 M134(xM117)=5 M117=5 |
| Han (NE China) | 0.524 | 42 | Katoh 2005 | M122=22 |
| Han (Meixian) | 0.514 | 35 | Xue 2006 | M122(xM159, M7, M134)=10 M117=4 M7=2 M159=1 M134(xM117)=1 |
| CHB (Han Chinese in Beijing) | 0.500 | 46 | Poznik 2016 | F444=8 M117=7 JST002611=5 KL2(xJST002611)=2 M188(xM7)=1 |
| Han (South China) | 0.492 | 65 | Yan 2011 | M122 |
| Va | 0.48 | Shi 2009 | ||
| Bai | 0.48 | Shi 2009 Wen 2004 | ||
| KHV (Kinh in Ho Chi Minh City) | 0.478 | 46 | Poznik 2016 | M7=6 M133=4 F444=4 JST002611=4 KL2(xJST002611)=2 N6>F4124=1 CTS1754=1 |
| Koreans | 0.472 | 216 | Kim 2007 | |
| Lisu | 0.47 | Shi 2009 | ||
| Hani | 0.47 | Wen 2004 | ||
| Han (Yili) | 0.469 | 32 | Xue 2006 | M122(xM159, M7, M134)=10 M7=2 M117=2 M134(xM117)=1 |
| Bai (Dali, Yunnan) | 0.46 | 50 | Wen 2004 | M122 |
| Mongols (Baotou) | 0.455 | 33 | F273=2 F4249=2 FGC23868=1 Z26109=1 F133=1 F12=1 Y26383=1 CTS201=1 F8=1 Y20928=1 F748=1 SK1783=1 SK1775=1 | |
| Hezhe (China) | 0.444 | 45 | Xue 2006 | M122(xM159, M7, M134)=11 M134(xM117)=2 M117=7 |
| Koreans | 0.443 | 506 | Kim 2011 | P201=146 M324(xP201)=76 M122(xM324)=2 |
| Tibetans (Zhongdian, Yunnan) | 0.440 | 50 | Wen 2004 | M122 |
| Miao | 0.44 | Shi 2009 | ||
| Yi | 0.44 | Wen 2004 | ||
| Lahu | 0.43 | Shi 2009 | ||
| Bit (Laos) | 0.429 | 28 | Cai 2011 | M122 |
| Manchu (NE China) | 0.426 | 101 | Katoh 2005 | M122=43 |
| Koreans (Seoul) | 0.422 | 573 | Park 2012 | M122 |
| Koreans (Daejeon) | 0.414 | 133 | Park 2012 | M122 |
| Hmong Daw (Laos) | 0.412 | 51 | Cai 2011 | M122 |
| Vietnamese | 0.41 | Karafet 2001 | ||
| Dai | 0.4 | Yang 2005 | ||
| Dungan (Kyrgyzstan) | 0.40 | 40 | Wells 2001 | M122 |
| Tibetans | 0.400 | 35 | Xue 2006 | M117=13 M134(xM117)=1 |
| Koreans (China) | 0.400 | 25 | Xue 2006 | M122(xM159, M7, M134)=6 M117=4 |
| Shan (Northern Thailand) | 0.400 | 20 | Brunelli 2017 | M117=7 M7=1 |
| Thai (Central Thailand) | 0.395 | 129 | Kutanan 2019 | F8/F42*=17 M7=11 JST002611=10 F474/F317=4 F323/F46=4 M159=2 F2055/CTS445=1 F2137=1 F837=1 |
| Koreans (South Korea) | 0.395 | 43 | Xue 2006 | M122(xM159, M7, M134)=7 M134(xM117)=5 M117=5 |
| Vietnamese | 0.39 | Jin 2009 | ||
| Khon Mueang (Northern Thailand) | 0.390 | 205 | Brunelli 2017 | O-M117=46 O-M7=17 O-M324(xM7, M134)=16 O-M122(xM324)=1 |
| Mon (Northern Thailand) | 0.389 | 18 | Brunelli 2017 | M117=4 M324(xM7, M134)=3 |
| Blang (Yunnan) | 0.385 | 52 | Cai 2011 | M122 |
| Northern Thai people (Khon Mueang & Tai Yuan) | 0.384 | 86 | Kutanan 2019 | F8/F42=24 M7=7 JST002611=1 F999/F717=1 |
| Manchu | 0.38 | Karafet 2001 | ||
| Philippine (Tagalog language group) | 0.380 | 50 | Tajima 2004 | M122 |
| Hanoi, Vietnam | 0.375 | 24 | Trejaut 2014 | M7=3 M134(xM133)=3 M133=1 JST002611=1 M159=1 |
| Manchu | 0.371 | 35 | Xue 2006 | M122(xM159, M7, M134)=6 M117=5 M134(xM117)=2 |
| Han (Lanzhou) | 0.367 | 30 | Xue 2006 | M122(xM159, M7, M134)=6 M117=3 M134(xM117)=2 |
| Lahu | 0.36 | Wen 2004 | ||
| Qiang | 0.364 | 33 | Xue 2006 | M134(xM117)=4 M117=3 M122(xM159, M7, M134)=3 M7=2 |
| Bamar (Myanmar) | 0.361 | 72 | Page23=26 | |
| Borneo, Indonesia | 0.360 | 86 | Karafet 2010 | M122 |
| Korean | 0.356 | 45 | Wells 2001 | M122 |
| Pahng (Guangxi) | 0.355 | 31 | Cai 2011 | M122 |
| Philippines | 0.354 | 48 | Karafet 2010 | M122 |
| Western Yugur | 0.35 | Zhou 2008 | ||
| Thai (Chiang Mai & Khon Kaen) | 0.353 | 34 | Shi 2009 Tajima 2004 | M122 |
| Tai Yong (Northern Thailand) | 0.346 | 26 | Brunelli 2017 | M324(xM7, M134)=4 M117=3 M7=2 |
| Tharu | 0.345 | 171 | Fornarino 2009 | M134 |
| Kinh (Hanoi, Vietnam) | 0.342 | 76 | He 2012 | M122 |
| Koreans (Seoul) | 0.341 | 85 | Katoh 2005 | M122=29 |
| Tibet | 0.340 | 156 | Gayden 2007 | M122 |
| Yao (Bama) | 0.343 | 35 | Xue 2006 | M7=12 |
| Kazakhs (SE Altai) | 0.337 | 89 | Dulik 2011 | M134(xM117, P101) |
| Tai Yuan (Thailand) | 0.329 | 85 | Brunelli 2017 | M117=15 M7=5 M122(xM324)=5 M134(xM117)=3 |
| Dai (Xishuangbanna, Yunnan) | 0.327 | 52 | Poznik 2016 | O-M133=13 O-M7=2 O-F444=1 O-JST002611=1 |
| Polynesians | 0.325 | Su 2000 | ||
| Tibetans | 0.32 | Wen 2004 | ||
| Khasi | 0.32 | Reddy 2007 | ||
| Lao (Luang Prabang, Laos) | 0.32 | 25 | He 2012 | M122 |
| Eastern Yugur | 0.31 | Zhou 2008 | ||
| Malays | 0.31 | Karafet 2001 | ||
| Buyei | 0.314 | 35 | Xue 2006 | M7=6 M134(xM117)=3 M117=1 M122(xM159, M7, M134)=1 |
| Mongolian (Khalkh) | 0.311 | Kim 2011 | ||
| Filipinos | 0.308 | 146 | Trejaut 2014 | P164(xM134)=26 JST002611=7 M7=3 M133=3 M134(xM133)=2 P201(xM159, M7, P164)=2 M159=1 M324(xKL1, P201)=1 |
| Han (Pinghua speakers) | 0.3 | Gan 2008 | ||
| Salar | 0.302 | 43 | Wang 2003 | M122 |
| Dong | 0.300 | 20 | Xie 2004 | M134=3 M122(xM7, M134)=3 |
| Thailand | 0.293 | 75 | Trejaut 2014 | M133(xM162)=10 M7=5 M134(xM133)=3 JST002611=2 P164(xM134)=1 M122(xM324)=1 |
| Koreans (NE China) | 0.291 | 79 | Katoh 2005 | M122=23 |
| Khasi | 0.29 | Kumar 2007 | ||
| Zhuang | 0.29 | Su 2000 | ||
| Inner Mongolian | 0.289 | 45 | Xue 2006 | M122(xM159, M7, M134)=5 M117=5 M134(xM117)=3 |
| Tai Lue (Northern Thailand) | 0.286 | 91 | Brunelli 2017 | O-M117=16 O-M7=6 O-M324(xM7, M134)=3 O-M122(xM324)=1 |
| Zhuang | 0.286 | 28 | Xie 2004 | M134=7 M122(xM7, M134)=1 |
| Laotian (Vientiane & Luang Prabang) | 0.275 | 40 | Kutanan 2019 | F8/F42=6 M7=2 M188(xM7)=2 P164(xF8,F46,F4110,F706,F717)=1 |
| Bonan | 0.273 | 44 | Wang 2003 | M122 |
| Sibe | 0.268 | 41 | Xue 2006 | M134(xM117)=5 M122(xM159, M7, M134)=4 M117=2 |
| Micronesia | 0.27 | Su 2000 | ||
| Mon (Thailand) | 0.267 | 105 | Kutanan 2019 | F8/F42=15 M7=4 F323/F46=4 JST002611=3 F2859=1 M122(x002611,M188,P164,F837)=1 |
| Daur | 0.256 | 39 | Xue 2006 | M122(xM159, M7, M134)=7 M117=3 |
| Polynesians | 0.25 | Hammer 2005 Kayser 2006 | ||
| Bunu (Guangxi) | 0.25 | 36 | Cai 2011 | M122 |
| Malay (near Kuala Lumpur) | 0.25 | 12 | Tajima 2004 | M122 |
| Zhuang (Guangxi) | 0.247 | 166 | Chen 2006 | M122(xM121, M134)=23 M117=9 M134(xM117)=7 M121=2 |
| Japanese (Kyūshū) | 0.240 | 104 | Tajima 2004 | M122 |
| Dongxiang | 0.24 | Wang 2003 | ||
| Manchurian Evenks | 0.24 | Karafet 2001 | ||
| Thai (Northern Thailand) | 0.235 | 17 | He 2012 | M122 |
| Japanese (Kagawa) | 0.234 | 47 | Xue 2006 | M117=8 M134(xM117)=2 M122(xM159, M7, M134)=1 |
| Mosuo (Ninglang, Yunnan) | 0.234 | 47 | Wen 2004 | M122 |
| Evenks (China) | 0.231 | 26 | Xue 2006 | M117=4 M134(xM117)=1 M122(xM159, M7, M134)=1 |
| Mongolia (mainly Khalkhs[84]) | 0.228 | 149 | Hammer 2005 | M134=24 M122(xM134)=10 |
| Zhuang (Napo County, Guangxi) | 0.222 | 63 | M117=5 M122(xM188, M134)=4 M188=3 M134(xM117)=2 | |
| Lawa (Northern Thailand) | 0.220 | 50 | Brunelli 2017 | M324(xM7, M134)=6 M117=5 |
| Mal (Laos) | 0.220 | 50 | Cai 2011 | M122 |
| Cambodian (Siem Reap) | 0.216 | 125 | Black 2006 | M122 |
| Japanese (Tokushima) | 0.214 | 70 | Hammer 2005 | M134=11 M122(xM134, LINE)=2 LINE=2 |
| Newar | 0.212 | 66 | Gayden 2007 | M117 |
| Lao Isan | 0.210 | 62 | Kutanan 2019 | M7=6 F8=4 JST002611=3 |
| Blang | 0.21 | Shi 2009 | ||
| Okinawans | 0.21 | Nonaka 2007 | ||
| Tai Khün (Northern Thailand) | 0.208 | 24 | Brunelli 2017 | M117=4 M134(xM117)=1 |
| Kathmandu, Nepal | 0.208 | 77 | Gayden 2007 | M324 |
| Sui | 0.200 | 50 | Xie 2004 | M134=10 |
| Yi (Shuangbai, Yunnan) | 0.20 | 50 | Wen 2004 | M122(xM7) |
| Japanese (Shizuoka) | 0.197 | 61 | Hammer 2005 | M122(xM134, LINE)=7 M134=5 |
| Khmu (Laos) | 0.196 | 51 | Cai 2011 | M122 |
| Dongxiang | 0.196 | 46 | Wang 2003 | M122 |
| Oroqen | 0.194 | 31 | Xue 2006 | M122(xM159, M7, M134)=2 M7=2 M134(xM117)=1 M117=1 |
| Khalkh (Mongolia) | 0.188 | 85 | Katoh 2005 | M122=16 |
| Japanese (Miyazaki) | 0.183 | 1285 | Nohara 2021 | M134=118 M122(xM134)=117 |
| Japanese (Tokyo) | 0.179 | 56 | Poznik 2016 | M117=5 M134(xM117)=3 JST002611=2 |
| Hani | 0.176 | 34 | Xue 2006 | M134(xM117)=3 M117=2 M122(xM159, M7, M134)=1 |
| Micronesia | 0.176 | 17 | Hammer 2005 | M122(xM134, LINE)=3 |
| Hui | 0.171 | 35 | Xue 2006 | M122(xM159, M7, M134)=4 M134(xM117)=1 M117=1 |
| Kalmyk (Khoshuud) | 0.171 | 82 | Malyarchuk 2013 | M122=14 |
| Japanese | 0.167 | 263 | Nonaka 2007 | M122 |
| Mandar (Sulawesi) | 0.167 | 54 | Karafet 2010 | M122 |
| Mulam (Luocheng) | 0.167 | 42 | Wang 2003 | JST002611=3 M134(xM117)=3 M117=1 |
| Japanese (Kantō) | 0.162 | 117 | Katoh 2005 | M122=19 |
| Thai | 0.16 | Jin 2009 | ||
| Zhuang | 0.16 | Karafet 2001 | ||
| Aheu (Laos) | 0.158 | 38 | Cai 2011 | M122 |
| Bugan (Yunnan) | 0.156 | 32 | Cai 2011 | M122 |
| Okinawans | 0.156 | 45 | Hammer 2005 | M122(xM134, LINE)=3 LINE=3 M134=1 |
| Uygur (Yili) | 0.154 | 39 | Xue 2006 | M122(xM159, M7, M134)=2 M134(xM117)=2 M117=2 |
| Japanese (Aomori) | 0.154 | 26 | Hammer 2005 | M134=3 M122(xM134, LINE)=1 |
| Cambodia | 0.14 | Shi 2009 | ||
| Cham (Binh Thuan, Vietnam) | 0.136 | 59 | He 2012 | M122 |
| Java (mainly sampled in Dieng) | 0.131 | 61 | Karafet 2010 | M122 |
| Aboriginal Taiwanese | 0.126 | 223 | Tajima 2004 | M122 |
| Uighur (Kazakhstan) | 0.122 | 41 | Wells 2001 | M122 |
| Uzbek (Bukhara) | 0.121 | 58 | Wells 2001 | M122 |
| Ulchi | 0.115 | 52 | O-M122(xP201)=6 | |
| Karakalpak (Uzbekistan) | 0.114 | 44 | Wells 2001 | M122 |
| Utsat (Sanya, Hainan) | 0.111 | 72 | Li 2013 | M117=3 M122(xM159, M117)=3 M159=2 |
| Outer Mongolian | 0.108 | 65 | Xue 2006 | M122(xM159, M7, M134)=3 M117=3 M134(xM117)=1 |
| Bo (Laos) | 0.107 | 28 | Cai 2011 | M122 |
| Tibetans | 0.1 | Zhou 2008 | ||
| Maluku Islands | 0.1 | 30 | Karafet 2010 | M122 |
| Kazakh (Kazakhstan) | 0.093 | 54 | Wells 2001 | M122 |
| Bouyei | 0.089 | 45 | Xie 2004 | M122(xM7, M134)=2 M7=1 M134=1 |
| Pumi (Ninglang, Yunnan) | 0.085 | 47 | Wen 2004 | M122(xM7) |
| Zakhchin (Mongolia) | 0.083 | 60 | Katoh 2005 | M122=5 |
| Mongols | 0.083 | 24 | Wells 2001 | M122 |
| Balinese (Bali) | 0.073 | 641 | Karafet 2010 | M122 |
| Japanese | 0.068 | 59 | Ochiai 2016 | P198 |
| Uriankhai (Mongolia) | 0.067 | 60 | Katoh 2005 | M122=4 |
| Sinte (Uzbekistan) | 0.067 | 15 | Wells 2001 | M122 |
| Uygur (Urumqi) | 0.065 | 31 | Xue 2006 | M134(xM117)=1 M117=1 |
| Iranian (Esfahan) | 0.063 | 16 | Wells 2001 | M122 |
| Kalmyk (Dörwöd) | 0.061 | 165 | Malyarchuk 2013 | M122=10 |
| Flores | 0.046 | 394 | Karafet 2010 | M122 |
| Buryat | 0.040 | 298 | Kharkov 2014 | M324(xM134)=5 M134(xM117)=4 M117=3 |
| Buyei | 0.04 | Yang 2005 | ||
| Kalmyk (Torguud) | 0.033 | 150 | Malyarchuk 2013 | M122=5 |
| Kazakhs (SW Altai) | 0.033 | 30 | Dulik 2011 | M134(xM117, P101) |
| Munda (Jharkhand) | 0.032 | 94 | M134=3 | |
| Burusho | 0.031 | 97 | Firasat 2007 | M122 |
| Li | 0.029 | 34 | Xue 2006 | M134(xM117)=1 |
| Sumba | 0.029 | 350 | Karafet 2010 | M122 |
| Khoton (Mongolia) | 0.025 | 40 | Katoh 2005 | M122=1 |
| Naxi (Lijiang, Yunnan) | 0.025 | 40 | Wen 2004 | M134 |
| Rajbanshi (West Bengal) | 0.022 | 45 | M134=1 | |
| Pathan | 0.010 | 96 | Firasat 2007 | M122 |
| Pakistan | 0.005 | 638 | Firasat 2007 | M122 |
Phylogenetics
Phylogenetic History
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O-M175 | 26 | VII | 1U | 28 | Eu16 | H9 | I | O* | O | O | O | O | O | O | O | O | O | O |
| O-M119 | 26 | VII | 1U | 32 | Eu16 | H9 | H | O1* | O1a | O1a | O1a | O1a | O1a | O1a | O1a | O1a | O1a | O1a |
| O-M101 | 26 | VII | 1U | 32 | Eu16 | H9 | H | O1a | O1a1 | O1a1a | O1a1a | O1a1 | O1a1 | O1a1a | O1a1a | O1a1a | O1a1a | O1a1a |
| O-M50 | 26 | VII | 1U | 32 | Eu16 | H10 | H | O1b | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 |
| O-P31 | 26 | VII | 1U | 33 | Eu16 | H5 | I | O2* | O2 | O2 | O2 | O2 | O2 | O2 | O2 | O2 | O2 | O2 |
| O-M95 | 26 | VII | 1U | 34 | Eu16 | H11 | G | O2a* | O2a | O2a | O2a | O2a | O2a | O2a | O2a | O2a | O2a1 | O2a1 |
| O-M88 | 26 | VII | 1U | 34 | Eu16 | H12 | G | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1a | O2a1a |
| O-SRY465 | 20 | VII | 1U | 35 | Eu16 | H5 | I | O2b* | O2b | O2b | O2b | O2b | O2b | O2b | O2b | O2b | O2b | O2b |
| O-47z | 5 | VII | 1U | 26 | Eu16 | H5 | I | O2b1 | O2b1a | O2b1 | O2b1 | O2b1a | O2b1a | O2b1 | O2b1 | O2b1 | O2b1 | O2b1 |
| O-M122 | 26 | VII | 1U | 29 | Eu16 | H6 | L | O3* | O3 | O3 | O3 | O3 | O3 | O3 | O3 | O3 | O3 | O3 |
| O-M121 | 26 | VII | 1U | 29 | Eu16 | H6 | L | O3a | O3a | O3a1 | O3a1 | O3a1 | O3a1 | O3a1 | O3a1 | O3a1 | O3a1a | O3a1a |
| O-M164 | 26 | VII | 1U | 29 | Eu16 | H6 | L | O3b | O3b | O3a2 | O3a2 | O3a2 | O3a2 | O3a2 | O3a2 | O3a2 | O3a1b | O3a1b |
| O-M159 | 13 | VII | 1U | 31 | Eu16 | H6 | L | O3c | O3c | O3a3a | O3a3a | O3a3 | O3a3 | O3a3a | O3a3a | O3a3a | O3a3a | O3a3a |
| O-M7 | 26 | VII | 1U | 29 | Eu16 | H7 | L | O3d* | O3c | O3a3b | O3a3b | O3a4 | O3a4 | O3a3b | O3a3b | O3a3b | O3a2b | O3a2b |
| O-M113 | 26 | VII | 1U | 29 | Eu16 | H7 | L | O3d1 | O3c1 | O3a3b1 | O3a3b1 | - | O3a4a | O3a3b1 | O3a3b1 | O3a3b1 | O3a2b1 | O3a2b1 |
| O-M134 | 26 | VII | 1U | 30 | Eu16 | H8 | L | O3e* | O3d | O3a3c | O3a3c | O3a5 | O3a5 | O3a3c | O3a3c | O3a3c | O3a2c1 | O3a2c1 |
| O-M117 | 26 | VII | 1U | 30 | Eu16 | H8 | L | O3e1* | O3d1 | O3a3c1 | O3a3c1 | O3a5a | O3a5a | O3a3c1 | O3a3c1 | O3a3c1 | O3a2c1a | O3a2c1a |
| O-M162 | 26 | VII | 1U | 30 | Eu16 | H8 | L | O3e1a | O3d1a | O3a3c1a | O3a3c1a | O3a5a1 | O3a5a1 | O3a3c1a | O3a3c1a | O3a3c1a | O3a2c1a1 | O3a2c1a1 |
Original Research Publications
The following research teams per their publications were represented in the creation of the YCC Tree.
Phylogenetic Trees
This phylogenetic tree of haplogroup O subclades is based on the YCC 2008 tree [85] and subsequent published research.
- O-M122 (M122, P198)
- O-P93 (M324, P93, P197, P198, P199, P200)
- O-M121 (M121, P27.2)
- O-M164 (M164)
- O-P201 (P201/021354)
- O-002611 (002611)
- O-M300 (M300)
- O-M333 (M333)
- O-P93 (M324, P93, P197, P198, P199, P200)
See also
Genetics
Y-DNA O Subclades
Y-DNA Backbone Tree
References
Citations
- 1 2 Karmin, Monika; Flores, Rodrigo; Saag, Lauri; et al. (28 February 2022). "Episodes of Diversification and Isolation in Island Southeast Asian and Near Oceanian Male Lineages". Molecular Biology and Evolution. 39 (3). doi:10.1093/molbev/msac045. ISSN 0737-4038. PMC 8926390. PMID 35294555.
- 1 2 Poznik, G. David; Xue, Yali; Mendez, Fernando L.; et al. (June 2016). "Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences". Nature Genetics. 48 (6): 593–599. doi:10.1038/ng.3559. PMC 4884158. PMID 27111036.
- 1 2 3 Shi 2009.
- ↑ Krahn and FTDNA 2013.
- 1 2 3 4 Cordaux, R. (12 February 2004). "The Northeast Indian Passageway: A Barrier or Corridor for Human Migrations?". Molecular Biology and Evolution. 21 (8): 1525–1533. doi:10.1093/molbev/msh151. ISSN 0737-4038. PMID 15128876.
- ↑ Gayden, Tenzin; Cadenas, Alicia M.; Regueiro, Maria; et al. (May 2007). "The Himalayas as a Directional Barrier to Gene Flow". The American Journal of Human Genetics. 80 (5): 884–894. doi:10.1086/516757. ISSN 0002-9297. PMC 1852741. PMID 17436243.
- 1 2 Bing Su, Chunjie Xiao, Ranjan Deka, Mark T. Seielstad, Daoroong Kangwanpong, Junhua Xiao, Daru Lu, Peter Underhill, Luca Cavalli-Sforza, Ranajit Chakraborty, Li Jin, "Y chromosome haplotypes reveal prehistorical migrations to the Himalayas." Hum. Genet. (2000) 107:582-590. DOI 10.1007/s004390000406 https://www.researchgate.net/publication/225570045_Y_chromosome_haplotypes_reveal_prehistorical_migrations_to_the_Himalayas
- 1 2 3 Ashirbekov, E. E.; Botbaev, D. M.; Belkozhaev, A. M.; Abayldaev, A. O.; Neupokoeva, A. S.; Mukhataev, J. E.; Alzhanuly, B.; Sharafutdinova, D. A.; Mukushkina, D. D.; Rakhymgozhin, M. B.; Khanseitova, A. K.; Limborska, S. A.; Aytkhozhina, N. A. "Distribution of Y-Chromosome Haplogroups of the Kazakh from the South Kazakhstan, Zhambyl, and Almaty Regions". Reports of the National Academy of Sciences of the Republic of Kazakhstan. 6 (316): 85–95. ISSN 2224-5227.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 Cai X, Qin Z, Wen B, Xu S, Wang Y, et al. (2011), "Human Migration through Bottlenecks from Southeast Asia into East Asia during Last Glacial Maximum Revealed by Y Chromosomes." PLoS ONE 6(8): e24282. doi:10.1371/journal.pone.0024282
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 Tatiana M. Karafet, Brian Hallmark, Murray P. Cox, et al. (2010), "Major East–West Division Underlies Y Chromosome Stratification across Indonesia." Mol. Biol. Evol. 27(8):1833–1844. doi:10.1093/molbev/msq063
- ↑ Wibhu Kutanan, Rasmi Shoocongdej, Metawee Srikummool, et al. (2020), "Cultural variation impacts paternal and maternal genetic lineages of the Hmong-Mien and Sino-Tibetan groups from Thailand." European Journal of Human Genetics https://doi.org/10.1038/s41431-020-0693-x
- 1 2 3 4 5 Enrico Macholdt, Leonardo Arias, Nguyen Thuy Duong, et al., "The paternal and maternal genetic history of Vietnamese populations." European Journal of Human Genetics (2020) 28:636–645. https://doi.org/10.1038/s41431-019-0557-4
- 1 2 Jatupol Kampuansai, Wibhu Kutanan, Eszter Dudás, Andrea Vágó-Zalán, Anikó Galambos, and Horolma Pamjav (2020), "Paternal genetic history of the Yong population in northern Thailand revealed by Y-chromosomal haplotypes and haplogroups." Molecular Genetics and Genomics volume 295, pages 579–589. https://doi.org/10.1007/s00438-019-01644-x
- 1 2 3 4 5 6 7 8 9 10 11 Kutanan, Wibhu; Kampuansai, Jatupol; Srikummool, Metawee; Brunelli, Andrea; Ghirotto, Silvia; Arias, Leonardo; Macholdt, Enrico; Hübner, Alexander; Schröder, Roland; Stoneking, Mark (2019). "Contrasting Paternal and Maternal Genetic Histories of Thai and Lao Populations". Mol. Biol. Evol. 36 (7): 1490–1506. doi:10.1093/molbev/msz083. PMC 6573475. PMID 30980085.
- 1 2 3 4 5 Brunelli, A; Kampuansai, J; Seielstad, M; Lomthaisong, K; Kangwanpong, D; Ghirotto, S; et al. (2017). "Y chromosomal evidence on the origin of northern Thai people". PLOS ONE. 12 (7): e0181935. Bibcode:2017PLoSO..1281935B. doi:10.1371/journal.pone.0181935. PMC 5524406. PMID 28742125.
- ↑ "O-M122单倍群详情".
- ↑ Gan 2008.
- ↑ Wen 2004c.
- 1 2 3 4 5 Hammer 2005.
- ↑ Hurles 2005.
- 1 2 3 4 Su 2000.
- ↑ Zhou 2008.
- 1 2 3 4 Wen 2004.
- ↑ Karafet 2001.
- ↑ Jing 2006.
- ↑ Li 2008.
- ↑ Katoh 2005.
- 1 2 Nonaka 2007.
- 1 2 3 4 Wells 2001.
- 1 2 3 Wang 2003.
- ↑ Yamamoto; et al. (2013). "Y-chromosome lineage in five regional Mongolian populations". Forensic Science International. Genetics Supplement Series 4: e260–e261. doi:10.1016/j.fsigss.2013.10.133 – via Elsevier Science Direct.
- ↑ Malyarchuk, Boris; Derenko, Miroslava; Denisova, Galina; Khoyt, Sanj; Woźniak, Marcin; Grzybowski, Tomasz; Zakharov, Ilya (2013). "Y-chromosome diversity in the Kalmyks at the ethnical and tribal levels". Journal of Human Genetics. 58 (12): 804–811. doi:10.1038/jhg.2013.108. PMID 24132124.
- ↑ Kharkov 2007.
- ↑ Di Cristofaro, J; Pennarun, E; Mazières, S; Myres, NM; Lin, AA; et al. (2013). "Afghan Hindu Kush: Where Eurasian Sub-Continent Gene Flows Converge". PLOS ONE. 8 (10): e76748. Bibcode:2013PLoSO...876748D. doi:10.1371/journal.pone.0076748. PMC 3799995. PMID 24204668.
- ↑ Kharkov, V. N.; Khamina, K. V.; Medvedeva, O. F.; Simonova, K. V.; Eremina, E. R.; Stepanov, V. A. (2014). "Gene pool of Buryats: Clinal variability and territorial subdivision based on data of Y-chromosome markers". Russian Journal of Genetics. 50 (2): 180–190. doi:10.1134/S1022795413110082. S2CID 15595963.
- ↑ Zhao, Yong-Bin; Zhang, Ye; Zhang, Quan-Chao; Li, Hong-Jie; Cui, Ying-Qiu; Xu, Zhi; Jin, Li; Zhou, Hui; Zhu, Hong (May 4, 2015). "Ancient DNA Reveals That the Genetic Structure of the Northern Han Chinese Was Shaped Prior to 3,000 Years Ago". PLOS ONE. 10 (5): e0125676. Bibcode:2015PLoSO..1025676Z. doi:10.1371/journal.pone.0125676. PMC 4418768. PMID 25938511.
- ↑ Yan, Shi; Wang, Chuan-Chao; Zheng, Hong-Xiang; Wang, Wei; Qin, Zhen-Dong; Wei, Lan-Hai; Wang, Yi; Pan, Xue-Dong; Fu, Wen-Qing; He, Yun-Gang; Xiong, Li-Jun; Jin, Wen-Fei; Li, Shi-Lin; An, Yu; Li, Hui; Jin, Li (August 29, 2014). "Y Chromosomes of 40% Chinese Descend from Three Neolithic Super-Grandfathers". PLOS ONE. 9 (8): e105691. arXiv:1310.3897. Bibcode:2014PLoSO...9j5691Y. doi:10.1371/journal.pone.0105691. PMC 4149484. PMID 25170956.
- ↑ Yao, Hong-Bing; Wang, Chuan-Chao; Tao, Xiaolan; Shang, Lei; Wen, Shao-Qing; Zhu, Bofeng; Kang, Longli; Jin, Li; Li, Hui (2016). "Genetic evidence for an East Asian origin of Chinese Muslim populations Dongxiang and Hui". Scientific Reports. 6: 38656. Bibcode:2016NatSR...638656Y. doi:10.1038/srep38656. PMC 5141421. PMID 27924949.
- 1 2 Reddy 2007.
- 1 2 Gayden 2007.
- 1 2 Fornarino, Simona; Pala, Maria; Battaglia, Vincenza; et al. (2009). "Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation". BMC Evolutionary Biology. 2009 (9): 154. Bibcode:2009BMCEE...9..154F. doi:10.1186/1471-2148-9-154. PMC 2720951. PMID 19573232.
- ↑ Murray P. Cox (June 2003). Genetic Patterning at Austronesian Contact Zones (PDF) (PhD). University of Otago. Archived from the original (PDF) on 5 September 2006. Retrieved 15 January 2022.
- ↑ Karafet 2005.
- 1 2 3 4 5 6 7 Karafet 2010.
- 1 2 3 Shi, Hong; Dong, Yong-li; Wen, Bo; Xiao, Chun-Jie; Underhill, Peter A.; Shen, Pei-dong; Chakraborty, Ranajit; Jin, Li; Su, Bing (Sep 2005). "Y-Chromosome Evidence of Southern Origin of the East Asian–Specific Haplogroup O2-M122". American Journal of Human Genetics. 77 (408–419): 408–19. doi:10.1086/444436. PMC 1226206. PMID 16080116.
- ↑ Peng, Min-Sheng; He, Jun-Dong; Fan, Long; et al. (2013). "Retrieving Y chromosomal haplogroup trees using GWAS data". European Journal of Human Genetics. 2013 (8): 1–5. doi:10.1038/ejhg.2013.272. PMC 4350590. PMID 24281365.
- ↑ Kim, Soon-Hee; Kim, Ki-Cheol; Shin, Dong-Jik; Jin, Han-Jun; Kwak, Kyoung-Don; Han, Myun-Soo; Song, Joon-Myong; Kim, Won; Kim, Wook (2011). "High frequencies of Y-chromosome haplogroup O2b-SRY465 lineages in Korea: a genetic perspective on the peopling of Korea". Investigative Genetics. 2 (1): 10–121. doi:10.1186/2041-2223-2-10. PMC 3087676. PMID 21463511.
- 1 2 3 4 5 Jin Park, Myung; Young Lee, Hwan; Ick Yang, Woo; Shin, Kyoung-Jin (2012). "Understanding the Y chromosome variation in Korea—relevance of combined haplogroup and haplotype analyses". International Journal of Legal Medicine. 126 (4): 589–599. doi:10.1007/s00414-012-0703-9. PMID 22569803. S2CID 27644576.
- 1 2 He 2012.
- 1 2 Trejaut, Jean A; Poloni, Estella S; Yen, Ju-Chen; Lai, Ying-Hui; Loo, Jun-Hun; Lee, Chien-Liang; He, Chun-Lin; Lin, Marie (2014). "Taiwan Y-chromosomal DNA variation and its relationship with Island Southeast Asia". BMC Genetics. 2014 (15): 77. doi:10.1186/1471-2156-15-77. PMC 4083334. PMID 24965575.
- 1 2 3 Underhill, Peter A.; Shen, Peidong; Lin, Alice A.; et al. (November 2000). "Y chromosome sequence variation and the history of human populations". Nature Genetics. 26 (3): 358–61. doi:10.1038/81685. PMID 11062480. S2CID 12893406.
- 1 2 Yan 2011.
- ↑ ISOGG Y-DNA Haplogroup O and its Subclades - 2017
- 1 2 YFull Haplogroup YTree v6.01 at 4 January 2018
- 1 2 3 4 5 6 Phylogenetic tree of human Y-DNA at 23mofang
- ↑ Wang, Chuan-Chao; Wang, Ling-Xiang; Shrestha, Rukesh; Zhang, Manfei; Huang, Xiu-Yuan; Hu, Kang; Jin, Li; Li, Hui (2014). "Genetic Structure of Qiangic Populations Residing in the Western Sichuan Corridor". PLOS ONE. 9 (8): e103772. Bibcode:2014PLoSO...9j3772W. doi:10.1371/journal.pone.0103772. PMC 4121179. PMID 25090432.
- ↑ Wang; Yan, Shi; Dong, QIN; Lu, Yan; Qi; Liang, DING; Hai, WEI; Shi; Lin, LI; Ya; Jun, YANG; Jin, Li; Li, Hui (2013). "Late Neolithic expansion of ancient Chinese revealed by Y chromosome haplogroup O2a1c-JST002611". Journal of Systematics and Evolution. 51 (3): 280–286. doi:10.1111/j.1759-6831.2012.00244.x. S2CID 55081530.
- 1 2 Yeun Kwon, So; Young Lee, Hwan; Young Lee, Eun; Ick Yang, Woo; Shin, Kyoung-Jin (2015). "Confirmation of Y haplogroup tree topologies with newly suggested Y-SNPs for the C2, O2b and O3a subhaplogroups". Forensic Science International: Genetics. 19: 42–46. doi:10.1016/j.fsigen.2015.06.003. PMID 26103100.
- ↑ "O-Ims-Jst002611单倍群详情".
- 1 2 Shi Yan, Chuan-Chao Wang, Hui Li, Shi-Lin Li, Li Jin, and The Genographic Consortium, "An updated tree of Y-chromosome Haplogroup O and revised phylogenetic positions of mutations P164 and PK4." European Journal of Human Genetics (2011) 19, 1013–1015; doi:10.1038/ejhg.2011.64; published online 20 April 2011.
- 1 2 He, G., Wang, M., Miao, L. et al. "Multiple founding paternal lineages inferred from the newly-developed 639-plex Y-SNP panel suggested the complex admixture and migration history of Chinese people." Hum Genomics 17, 29 (2023). https://doi.org/10.1186/s40246-023-00476-6
- ↑ "O-M188单倍群详情".
- ↑ "O-Am01822单倍群详情".
- ↑ "O-M134单倍群详情".
- ↑ "O-M7单倍群详情".
- ↑ "O-Cts201单倍群详情".
- ↑ "O-Mf109044单倍群详情".
- ↑ "O-M134单倍群详情".
- ↑ "O-Am01822单倍群详情".
- 1 2 3 4 Xue 2006.
- ↑ "O-M159单倍群详情".
- ↑ O'Rourke, Dennis; Cai, Xiaoyun; Qin, Zhendong; Wen, Bo; Xu, Shuhua; Wang, Yi; Lu, Yan; Wei, Lanhai; Wang, Chuanchao; Li, Shilin; Huang, Xingqiu; Jin, Li; Li, Hui (2011). "Human Migration through Bottlenecks from Southeast Asia into East Asia during Last Glacial Maximum Revealed by Y Chromosomes". PLOS ONE. 6 (8): e24282. Bibcode:2011PLoSO...624282C. doi:10.1371/journal.pone.0024282. ISSN 1932-6203. PMC 3164178. PMID 21904623.
- 1 2 3 4 5 Cai 2011.
- 1 2 He J-D, Peng M-S, Quang HH, Dang KP, Trieu AV, et al. (2012), "Patrilineal Perspective on the Austronesian Diffusion in Mainland Southeast Asia." PLoS ONE 7(5): e36437. doi:10.1371/journal.pone.0036437
- 1 2 3 4 5 6 7 Manfred Kayser, Ying Choi, Mannis van Oven, et al. (2008), "The Impact of the Austronesian Expansion: Evidence from mtDNA and Y Chromosome Diversity in the Admiralty Islands of Melanesia." Mol. Biol. Evol. 25(7):1362–1374. doi:10.1093/molbev/msn078
- ↑ Frederick Delfin, Jazelyn M Salvador, Gayvelline C Calacal, et al., "The Y-chromosome landscape of the Philippines: extensive heterogeneity and varying genetic affinities of Negrito and non-Negrito groups." European Journal of Human Genetics (2011) 19, 224–230; doi:10.1038/ejhg.2010.162; published online 29 September 2010.
- 1 2 Zhang, X.; Tang, Z.; Wang, B.; Zhou, X.; Zhou, L.; Zhang, G.; Tian, J.; Zhao, Y.; Yao, Z.; Tian, L.; et al. "Forensic Analysis and Genetic Structure Construction of Chinese Chongming Island Han Based on Y Chromosome STRs and SNPs." Genes 2022, 13, 1363. https://doi.org/10.3390/genes13081363
- ↑ Karafet TM, Osipova LP, Savina OV, Hallmark B, Hammer MF. "Siberian genetic diversity reveals complex origins of the Samoyedic-speaking populations." Am J Hum Biol. 2018;e23194. https://doi.org/10.1002/ajhb.23194
- ↑ Myung Jin Park, Hwan Young Lee, Na Young Kim, et al. (2013), "Y-SNP miniplexes for East Asian Y-chromosomal haplogroup determination in degraded DNA." Forensic Science International: Genetics 7, 75–81. http://dx.doi.org/10.1016/j.fsigen.2012.06.014
- ↑ "夏商西南大族祖源分析-23魔方祖源基因检测".
- ↑ Nonaka, I.; Minaguchi, K.; Takezaki, N. (2007). "Y-chromosomal Binary Haplogroups in the Japanese Population and their Relationship to 16 Y-STR Polymorphisms". Annals of Human Genetics. 71 (4): 480–495. doi:10.1111/j.1469-1809.2006.00343.x. hdl:10130/491. PMID 17274803. S2CID 1041367.
- 1 2 Sae Naitoh, Iku Kasahara-Nonaka, Kiyoshi Minaguchi, and Phrabhakaran Nambiar, "Assignment of Y-chromosomal SNPs found in Japanese population to Y-chromosomal haplogroup tree." Journal of Human Genetics (2013) 58, 195–201; doi:10.1038/jhg.2012.159; published online 7 February 2013.
- ↑ "O-M117单倍群详情".
- ↑ Karafet, T. M.; Zegura, S. L.; Posukh, O.; Osipova, L.; Bergen, A.; Long, J.; Goldman, D.; Klitz, W.; Harihara, S.; de Knijff, P.; Wiebe, V.; Griffiths, R. C.; Templeton, A. R.; Hammer, M. F. (1999). "Ancestral Asian Source(s) of New World Y-Chromosome Founder Haplotypes". American Journal of Human Genetics. 64 (3): 817–831. doi:10.1086/302282. PMC 1377800. PMID 10053017.
- ↑ Karafet 2008.
Sources
- Journal articles
- Black, M. L.; Dufall, K.; Wise, C.; Sullivan, S.; Bittles, A. H. (2006). "Genetic ancestries in northwest Cambodia". Annals of Human Biology. 33 (5–6): 620–7. doi:10.1080/03014460600882561. PMID 17381059. S2CID 34579092.
- Brunelli, Andrea; Kampuansai, Jatupol; Seielstad, Mark; Lomthaisong, Khemika; Kangwanpong, Daoroong; Ghirotto, Silvia; Kutanan, Wibhu (24 July 2017). "Y chromosomal evidence on the origin of northern Thai people". PLOS ONE. 12 (7): e0181935. Bibcode:2017PLoSO..1281935B. doi:10.1371/journal.pone.0181935. PMC 5524406. PMID 28742125.
- Chen, Chen; Hui, LI; Zhen-Dong, QIN; Wen-Hong, LIU; Wei-Xiong, LIN; Rui-Xing, YIN; Li, JIN; Shang-Ling, PAN (2006). "Y-chromosome Genotyping and Genetic Structure of Zhuang Populations". Acta Genetica Sinica. 33 (12): 1060–72. CiteSeerX 10.1.1.602.5490. doi:10.1016/S0379-4172(06)60143-1. PMID 17185165.
- Cai, Xiaoyun; Qin, Zhendong; Wen, Bo; Xu, Shuhua; Wang, Yi; Lu, Yan; Wei, Lanhai; Wang, Chuanchao; et al. (2011). O'Rourke, Dennis (ed.). "Human Migration through Bottlenecks from Southeast Asia into East Asia during Last Glacial Maximum Revealed by Y Chromosomes". PLOS ONE. 6 (8): e24282. Bibcode:2011PLoSO...624282C. doi:10.1371/journal.pone.0024282. PMC 3164178. PMID 21904623.
- Cordaux, R.; Weiss, G; Saha, N; Stoneking, M (2004). "The Northeast Indian Passageway: A Barrier or Corridor for Human Migrations?". Molecular Biology and Evolution. 21 (8): 1525–33. doi:10.1093/molbev/msh151. PMID 15128876.
- Dulik, Matthew C.; Osipova, Ludmila P.; Schurr, Theodore G. (11 March 2011). "Y-Chromosome Variation in Altaian Kazakhs Reveals a Common Paternal Gene Pool for Kazakhs and the Influence of Mongolian Expansions". PLOS ONE. 6 (3): e17548. Bibcode:2011PLoSO...617548D. doi:10.1371/journal.pone.0017548. PMC 3055870. PMID 21412412.
- Firasat, Sadaf; Khaliq, Shagufta; Mohyuddin, Aisha; Papaioannou, Myrto; Tyler-Smith, Chris; Underhill, Peter A.; Ayub, Qasim (2007). "Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan". European Journal of Human Genetics. 15 (1): 121–126. doi:10.1038/sj.ejhg.5201726. PMC 2588664. PMID 17047675.
- Gan, Rui-Jing; Pan, Shang-Ling; Mustavich, Laura F.; Qin, Zhen-Dong; Cai, Xiao-Yun; Qian, Ji; Liu, Cheng-Wu; Peng, Jun-Hua; et al. (2008). "Pinghua population as an exception of Han Chinese's coherent genetic structure". Journal of Human Genetics. 53 (4): 303–13. doi:10.1007/s10038-008-0250-x. PMID 18270655.
- Gayden, Tenzin; Cadenas, Alicia M.; Regueiro, Maria; Singh, Nanda B.; Zhivotovsky, Lev A.; Underhill, Peter A.; Cavalli-Sforza, Luigi L.; Herrera, Rene J. (2007). "The Himalayas as a Directional Barrier to Gene Flow". The American Journal of Human Genetics. 80 (5): 884–94. doi:10.1086/516757. PMC 1852741. PMID 17436243.
- Hammer, Michael F.; Karafet, Tatiana M.; Park, Hwayong; Omoto, Keiichi; Harihara, Shinji; Stoneking, Mark; Horai, Satoshi (2005). "Dual origins of the Japanese: Common ground for hunter-gatherer and farmer Y chromosomes". Journal of Human Genetics. 51 (1): 47–58. doi:10.1007/s10038-005-0322-0. PMID 16328082.
- He, Jun-Dong; Peng, Min-Sheng; Quang, Huy Ho; Dang, Khoa Pham; Trieu, An Vu; Wu, Shi-Fang; Jin, Jie-Qiong; Murphy, Robert W.; et al. (2012). Kayser, Manfred (ed.). "Patrilineal Perspective on the Austronesian Diffusion in Mainland Southeast Asia". PLOS ONE. 7 (5): e36437. Bibcode:2012PLoSO...736437H. doi:10.1371/journal.pone.0036437. PMC 3346718. PMID 22586471.
- Hurles, M; Sykes, B; Jobling, M; Forster, P (2005). "The Dual Origin of the Malagasy in Island Southeast Asia and East Africa: Evidence from Maternal and Paternal Lineages". The American Journal of Human Genetics. 76 (5): 894–901. doi:10.1086/430051. PMC 1199379. PMID 15793703.
- Jin, Han-Jun; Tyler-Smith, Chris; Kim, Wook (2009). Batzer, Mark A (ed.). "The Peopling of Korea Revealed by Analyses of Mitochondrial DNA and Y-Chromosomal Markers". PLOS ONE. 4 (1): e4210. Bibcode:2009PLoSO...4.4210J. doi:10.1371/journal.pone.0004210. PMC 2615218. PMID 19148289.
- Jing, Chen; Hui, LI; Zhen-Dong, QIN; Wen-Hong, LIU; Wei-Xiong, LIN; Rui-Xing, YIN; Li, JIN; Shang-Ling, PAN (2006). "Y-chromosome Genotyping and Genetic Structure of Zhuang Populations". Acta Genetica Sinica. 33 (12): 1060–72. doi:10.1016/S0379-4172(06)60143-1. PMID 17185165.
- Karafet, Tatiana; Xu, Liping; Du, Ruofu; Wang, William; Feng, Shi; Wells, R.S.; Redd, Alan J.; Zegura, Stephen L.; Hammer, Michael F. (2001). "Paternal Population History of East Asia: Sources, Patterns, and Microevolutionary Processes". The American Journal of Human Genetics. 69 (3): 615–28. doi:10.1086/323299. PMC 1235490. PMID 11481588.
- Karafet, Tatiana M.; Lansing, J. S.; Redd, Alan J.; Watkins, Joseph C.; Surata, S. P. K.; Arthawiguna, W. A.; Mayer, Laura; Bamshad, Michael; et al. (2005). "Balinese Y-Chromosome Perspective on the Peopling of Indonesia: Genetic Contributions from Pre-Neolithic Hunter-Gatherers, Austronesian Farmers, and Indian Traders" (PDF). Human Biology. 77 (1): 93–114. doi:10.1353/hub.2005.0030. hdl:1808/13586. PMID 16114819. S2CID 7953854.
- Karafet, T. M.; Mendez, F. L.; Meilerman, M. B.; Underhill, P. A.; Zegura, S. L.; Hammer, M. F. (2008), "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree", Genome Research, 18 (5): 830–8, doi:10.1101/gr.7172008, PMC 2336805, PMID 18385274
- Karafet, T. M.; Hallmark, B.; Cox, M. P.; Sudoyo, H.; Downey, S.; Lansing, J. S.; Hammer, M. F. (2010). "Major East-West Division Underlies Y Chromosome Stratification across Indonesia". Molecular Biology and Evolution. 27 (8): 1833–44. doi:10.1093/molbev/msq063. PMID 20207712. S2CID 4819475.
- Katoh, Toru; Munkhbat, Batmunkh; Tounai, Kenichi; Mano, Shuhei; Ando, Harue; Oyungerel, Ganjuur; Chae, Gue-Tae; Han, Huun; Jia, Guan-Jun; Tokunaga, Katsushi; Munkhtuvshin, Namid; Tamiya, Gen; Inoko, Hidetoshi (2005). "Genetic features of Mongolian ethnic groups revealed by Y-chromosomal analysis". Gene. 346: 63–70. doi:10.1016/j.gene.2004.10.023. PMID 15716011.
- Kayser, M.; Brauer, S; Cordaux, R; Casto, A; Lao, O; Zhivotovsky, LA; Moyse-Faurie, C; Rutledge, RB; et al. (2006). "Melanesian and Asian Origins of Polynesians: MtDNA and Y Chromosome Gradients Across the Pacific". Molecular Biology and Evolution. 23 (11): 2234–44. doi:10.1093/molbev/msl093. hdl:11858/00-001M-0000-0010-0145-0. PMID 16923821.
- Kharkov, V. N.; Stepanov, V. A.; Medvedeva, O. F.; Spiridonova, M. G.; Voevoda, M. I.; Tadinova, V. N.; Puzyrev, V. P. (2007). "Gene pool differences between Northern and Southern Altaians inferred from the data on Y-chromosomal haplogroups". Russian Journal of Genetics. 43 (5): 551–562. doi:10.1134/S1022795407050110. PMID 17633562. S2CID 566825.
- Kim, Wook; Yoo, Tag-Keun; Kim, Sung-Joo; Shin, Dong-Jik; Tyler-Smith, Chris; Jin, Han-Jun; Kwak, Kyoung-Don; Kim, Eun-Tak; Bae, Yoon-Sun (2007). Blagosklonny, Mikhail (ed.). "Lack of Association between Y-Chromosomal Haplogroups and Prostate Cancer in the Korean Population". PLOS ONE. 2 (1): e172. Bibcode:2007PLoSO...2..172K. doi:10.1371/journal.pone.0000172. PMC 1766463. PMID 17245448.
- Kumar, Vikrant; Reddy, Arimanda NS; Babu, Jagedeesh P; Rao, Tipirisetti N; Langstieh, Banrida T; Thangaraj, Kumarasamy; Reddy, Alla G; Singh, Lalji; Reddy, Battini M (2007). "Y-chromosome evidence suggests a common paternal heritage of Austro-Asiatic populations". BMC Evolutionary Biology. 7 (1): 47. Bibcode:2007BMCEE...7...47K. doi:10.1186/1471-2148-7-47. PMC 1851701. PMID 17389048.
- Kutanan, Wibhu; Kampuansai, Jatupol; Srikummool, Metawee; Brunelli, Andrea; Ghirotto, Silvia; Arias, Leonardo; Macholdt, Enrico; Hübner, Alexander; Schröder, Roland; Stoneking, Mark (1 July 2019). "Contrasting Paternal and Maternal Genetic Histories of Thai and Lao Populations". Molecular Biology and Evolution. 36 (7): 1490–1506. doi:10.1093/molbev/msz083. PMC 6573475. PMID 30980085.</ref>
- Li, Hui; Wen, Bo; Chen, Shu-Juo; Su, Bing; Pramoonjago, Patcharin; Liu, Yangfan; Pan, Shangling; Qin, Zhendong; Liu, Wenhong; Cheng, Xu; Yang, Ningning; Li, Xin; Tran, Dinhbinh; Lu, Daru; Hsu, Mu-Tsu; Deka, Ranjan; Marzuki, Sangkot; Tan, Chia-Chen; Jin, Li (2008). "Paternal genetic affinity between western Austronesians and Daic populations". BMC Evolutionary Biology. 8 (1): 146. Bibcode:2008BMCEE...8..146L. doi:10.1186/1471-2148-8-146. PMC 2408594. PMID 18482451.
- Li, Dong-Na; Wang, Chuan-Chao; Yang, Kun; et al. (2013). "Substitution of Hainan Indigenous Genetic Lineage in the Utsat People, Exiles of the Champa Kingdom: Genetic Structure of Hainan Utsat People". Journal of Systematics and Evolution. 51 (3): 287–294. doi:10.1111/jse.12000.
- Nohara, Hirofumi; Maeda, Ikuko; Hisazumi, Rinnosuke; Uchiyama, Taketo; Hirashima, Hiroko; Nakata, Masahito; Ohno, Rika; Hasegawa, Tetsuro; Shimizu, Kenshi (2021). "Geographic distribution of Y-STR haplotypes and Y-haplogroups among Miyazaki Prefecture residents". Japanese Journal of Forensic Science and Technology. 26 (1): 17–27. doi:10.3408/jafst.778. S2CID 226748034.
- Nonaka, I.; Minaguchi, K.; Takezaki, N. (2007). "Y-chromosomal Binary Haplogroups in the Japanese Population and their Relationship to 16 Y-STR Polymorphisms". Annals of Human Genetics. 71 (4): 480–95. doi:10.1111/j.1469-1809.2006.00343.x. hdl:10130/491. PMID 17274803. S2CID 1041367.
- Ochiai E, Minaguchi K, Nambiar P, Kakimoto Y, Satoh F, Nakatome M, et al. (September 2016). "Evaluation of Y chromosomal SNP haplogrouping in the HID-Ion AmpliSeq™ Identity Panel". Legal Medicine. 22: 58–61. doi:10.1016/j.legalmed.2016.08.001. PMID 27591541.
- Park, Myung Jin; Lee, Hwan Young; Yang, Woo Ick; Shin, Kyoung-Jin (2012). "Understanding the Y chromosome variation in Korea—relevance of combined haplogroup and haplotype analyses". International Journal of Legal Medicine. 126 (4): 589–99. doi:10.1007/s00414-012-0703-9. PMID 22569803. S2CID 27644576.
- Poznik, G David; Xue, Yali; Mendez, Fernando L; Willems, Thomas F; Massaia, Andrea; Wilson Sayres, Melissa A; Ayub, Qasim; McCarthy, Shane A; et al. (June 2016). "Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences". Nature Genetics. 48 (6): 593–599. doi:10.1038/ng.3559. PMC 4884158. PMID 27111036.
- Reddy, B. Mohan; Langstieh, B. T.; Kumar, Vikrant; Nagaraja, T.; Reddy, A. N. S.; Meka, Aruna; Reddy, A. G.; Thangaraj, K.; Singh, Lalji (2007). Awadalla, Philip (ed.). "Austro-Asiatic Tribes of Northeast India Provide Hitherto Missing Genetic Link between South and Southeast Asia". PLOS ONE. 2 (11): e1141. Bibcode:2007PLoSO...2.1141R. doi:10.1371/journal.pone.0001141. PMC 2065843. PMID 17989774.
- Shi, Simona; Pala, Maria; Battaglia, Vincenza; Maranta, Ramona; Achilli, Alessandro; Modiano, Guido; Torroni, Antonio; Semino, Ornella; Santachiara-Benerecetti, Silvana A (2009). "Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): A reservoir of genetic variation". BMC Evolutionary Biology. 9 (1): 154. Bibcode:2009BMCEE...9..154F. doi:10.1186/1471-2148-9-154. PMC 2720951. PMID 19573232.
- Su, B.; Jin, L.; Underhill, P.; Martinson, J.; Saha, N.; McGarvey, S. T.; Shriver, M. D.; Chu, J.; et al. (2000). "Polynesian origins: Insights from the Y chromosome". Proceedings of the National Academy of Sciences. 97 (15): 8225–8228. Bibcode:2000PNAS...97.8225S. doi:10.1073/pnas.97.15.8225. PMC 26928. PMID 10899994.
*H6 (=O-M122(xO-M7, O-M134)) in 18/73=24.7% *H8 (=O-M134) in 2/73=2.7% for a total of 20/73=27.4% O-M122 in a pool of seven samples from Micronesia. *13/40=32.5% O-M122(xM7, M134) in a pool of three samples from Polynesia. *9/27=33.3% H6 (=O-M122(xM7, M134)) *6/27=22.2% H8 (=O-M134) for a total of 15/27=55.6% O-M122 in "Malay" sample *2/19=10.5% H6 (=O-M122(xM7, M134)) in "Kota Kinabalu" sample.
- Su, Bing; Xiao, Junhua; Underhill, Peter; Deka, Ranjan; Zhang, Weiling; Akey, Joshua; Huang, Wei; Shen, Di; et al. (1999). "Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age". The American Journal of Human Genetics. 65 (6): 1718–24. doi:10.1086/302680. PMC 1288383. PMID 10577926.
- Tajima, Atsushi; Hayami, Masanori; Tokunaga, Katsushi; Juji, Takeo; Matsuo, Masafumi; Marzuki, Sangkot; Omoto, Keiichi; Horai, Satoshi (2004). "Genetic origins of the Ainu inferred from combined DNA analyses of maternal and paternal lineages". Journal of Human Genetics. 49 (4): 187–93. doi:10.1007/s10038-004-0131-x. PMID 14997363.
- Wang, Wei; Wise, Cheryl; Baric, Tom; Black, Michael L.; Bittles, Alan H. (August 1, 2003). "The origins and genetic structure of three co-resident Chinese Muslim populations: the Salar, Bo'an and Dongxiang". Human Genetics. Springer-Verlag. 113 (3): 244–52. doi:10.1007/s00439-003-0948-y. ISSN 0340-6717. PMID 12759817. S2CID 11138499.
- Wells, R. S.; Yuldasheva, N.; Ruzibakiev, R.; Underhill, P. A.; Evseeva, I.; Blue-Smith, J.; Jin, L.; Su, B.; et al. (2001). "The Eurasian Heartland: A continental perspective on Y-chromosome diversity". Proceedings of the National Academy of Sciences. 98 (18): 10244–9. Bibcode:2001PNAS...9810244W. doi:10.1073/pnas.171305098. PMC 56946. PMID 11526236.
- Wen, Bo; Li, Hui; Lu, Daru; Song, Xiufeng; Zhang, Feng; He, Yungang; Li, Feng; Gao, Yang; et al. (2004). "Genetic evidence supports demic diffusion of Han culture". Nature. 431 (7006): 302–5. Bibcode:2004Natur.431..302W. doi:10.1038/nature02878. PMID 15372031. S2CID 4301581.
- Wen, Bo; Xie, Xuanhua; Gao, Song; Li, Hui; Shi, Hong; Song, Xiufeng; Qian, Tingzhi; Xiao, Chunjie; et al. (2004). "Analyses of Genetic Structure of Tibeto-Burman Populations Reveals Sex-Biased Admixture in Southern Tibeto-Burmans". The American Journal of Human Genetics. 74 (5): 856–865. doi:10.1086/386292. PMC 1181980. PMID 15042512.
- Wen, Bo; Hong, S; Ling, R; Huifeng, X; Kaiyuan, L; Wenyi, Z; Bing, S; Shiheng, S; et al. (2004). "The origin of Mosuo people as revealed by mtDNA and Y chromosome variation". Science China Life Sciences. 47 (1): 1–10. doi:10.1360/02yc0207. PMID 15382670. S2CID 7999778.
- Xie, Xuan-Hua (2004). "Genetic Structure of Tujia as Revealed by Y Chromosomes". Yi Chuan Xue Bao = Acta Genetica Sinica. 31 (10): 1023–9. PMID 15552034.
- Xue, Y.; Zerjal, T; Bao, W; Zhu, S; Shu, Q; Xu, J; Du, R; Fu, S; et al. (2006). "Male Demography in East Asia: A North-South Contrast in Human Population Expansion Times". Genetics. 172 (4): 2431–9. doi:10.1534/genetics.105.054270. PMC 1456369. PMID 16489223.
- Yan, Shi (September 2011). "An updated tree of Y-chromosome Haplogroup O and revised phylogenetic positions of mutations P164 and PK4". European Journal of Human Genetics. 19 (9): 1013–5. doi:10.1038/ejhg.2011.64. PMC 3179364. PMID 21505448.
- Yang, Zhili; Dong, Yongli; Gao, Lu; Cheng, Baowen; Yang, Jie; Zeng, Weimin; Lu, Jing; Su, Yanhua; Xiao, Chunjie (2005). "The distribution of Y chromosome haplogroups in the nationalities from Yunnan Province of China". Annals of Human Biology. 32 (1): 80–7. doi:10.1080/03014460400027557. PMID 15788357. S2CID 39153696.
- Zhou, Ruixia; Yang, Daqun; Zhang, Hua; Yu, Weiping; An, Lizhe; Wang, Xilong; Li, Hong; Xu, Jiujin; Xie, Xiaodong (2008). "Origin and evolution of two Yugur sub-clans in Northwest China: A case study in paternal genetic landscape". Annals of Human Biology. 35 (2): 198–211. doi:10.1080/03014460801922927. PMID 18428013. S2CID 5453358.
- Websites
- Genographic Project, R. Spencer; Wells. "The Genographic Project - Atlas of the Human Journey". Archived from the original on 2011-02-05.
- Krahn; FTDNA (2003). "Genomic Research Center Draft Tree (AKA Y-TRee)". Archived from the original on 2015-08-15.
Sources for conversion tables
- Capelli, Cristian; Wilson, James F.; Richards, Martin; Stumpf, Michael P.H.; et al. (February 2001). "A Predominantly Indigenous Paternal Heritage for the Austronesian-Speaking Peoples of Insular Southeast Asia and Oceania". The American Journal of Human Genetics. 68 (2): 432–443. doi:10.1086/318205. PMC 1235276. PMID 11170891.
- Hammer, Michael F.; Karafet, Tatiana M.; Redd, Alan J.; Jarjanazi, Hamdi; et al. (1 July 2001). "Hierarchical Patterns of Global Human Y-Chromosome Diversity". Molecular Biology and Evolution. 18 (7): 1189–1203. doi:10.1093/oxfordjournals.molbev.a003906. PMID 11420360.
- Jobling, Mark A.; Tyler-Smith, Chris (2000), "New uses for new haplotypes", Trends in Genetics, 16 (8): 356–62, doi:10.1016/S0168-9525(00)02057-6, PMID 10904265
- Kaladjieva, Luba; Calafell, Francesc; Jobling, Mark A; Angelicheva, Dora; et al. (February 2001). "Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages". European Journal of Human Genetics. 9 (2): 97–104. doi:10.1038/sj.ejhg.5200597. PMID 11313742. S2CID 21432405.
- Karafet, Tatiana; Xu, Liping; Du, Ruofu; Wang, William; et al. (September 2001). "Paternal Population History of East Asia: Sources, Patterns, and Microevolutionary Processes". The American Journal of Human Genetics. 69 (3): 615–628. doi:10.1086/323299. PMC 1235490. PMID 11481588.
- Semino, O.; Passarino, G; Oefner, PJ; Lin, AA; et al. (2000), "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective", Science, 290 (5494): 1155–9, Bibcode:2000Sci...290.1155S, doi:10.1126/science.290.5494.1155, PMID 11073453
- Su, Bing; Xiao, Junhua; Underhill, Peter; Deka, Ranjan; et al. (December 1999). "Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age". The American Journal of Human Genetics. 65 (6): 1718–1724. doi:10.1086/302680. PMC 1288383. PMID 10577926.
- Underhill, Peter A.; Shen, Peidong; Lin, Alice A.; Jin, Li; et al. (November 2000). "Y chromosome sequence variation and the history of human populations". Nature Genetics. 26 (3): 358–361. doi:10.1038/81685. PMID 11062480. S2CID 12893406.
Further reading
- Karmin, Monika; Saag, Lauri; Vicente, Mário; et al. (2015). "A recent bottleneck of Y chromosome diversity coincides with a global change in culture". Genome Research. 25 (4): 459–466. doi:10.1101/gr.186684.114. PMC 4381518. PMID 25770088.
External links
- Spread of Haplogroup O-M122, from The Genographic Project, National Geographic
- China DNA interest group at Facebook
- China DNA Project Website at Family Tree DNA