| Orthohepevirus A | |
|---|---|
 | |
| TEM micrograph of Orthohepevirus A virions | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Kitrinoviricota |
| Class: | Alsuviricetes |
| Order: | Hepelivirales |
| Family: | Hepeviridae |
| Genus: | Orthohepevirus |
| Species: | Orthohepevirus A |
| Synonyms[1] | |
| |
The hepatitis E virus (HEV) is the causative agent of hepatitis E. It is of the species Orthohepevirus A.[lower-alpha 1][2][1]
Globally, approximately 939 million corresponding to 1 in 8 individuals have ever experienced HEV infection. About 15–110 million individuals have recent or ongoing HEV infection.[3] The virus particle was first seen in 1983,[4] but was only molecularly cloned in 1989.[5]
Genome and proteome
Orthohepevirus A can be classified into eight different genotypes from different geographical regions: genotype 1 (Asia), genotype 2 (Africa and Mexico), genotype 3 (Europe and North America), genotype 4 (Asia); genotypes 5 and 6 have been detected in Asian wild boar and genotypes 7 and 8 in camels.[2][6]
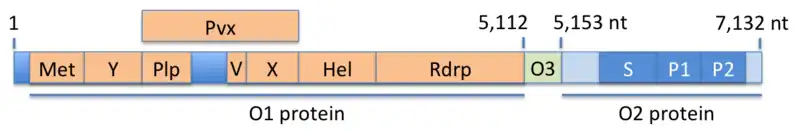
The viral genome is a single strand of positive-sense RNA that is about 7200 bases in length. The three open reading frames (ORF1, ORF2 and ORF3) encode for three proteins (O1, O2, O3), two of which are polyproteins, that is, they are cleaved into fragments which carry out the actual functions of the virus (see figure). The O1 protein consists of seven such fragments, namely Met (methyltransferase), Y (Y-domain), Plp (papain-like protease), V (proline-rich variable region), X (X-domain, macro-domain), Hel (helicase), and Rdrp (RNA-dependent RNA polymerase). The Pvx domain is a fusion protein consisting of the Plp, V, and X domains. The O3 protein is encoded by a single open-reading frame (ORF3). The O2 protein encodes the capsid, which is composed of three domains, namely the shell domain (S) and two protruding domains (P1, P2).[7] Numbers in the figure indicate positions in the RNA sequence.
Interactome
The protein-protein interactome among Orthohepevirus A proteins has been mapped by Osterman et al. (2015), who found 25 interactions among the 10 proteins studied. Almost all (24) of these interactions were considered as of "high quality".[8]
Structure
The viral particles are 27 to 34 nanometers in diameter and are not enveloped.[2][4]
Taxonomy
It was previously classified in the family Caliciviridae. However, its genome more closely resembles rubella virus. It is now classified as a member of the genus Orthohepevirus in the family Hepeviridae.[2]
Evolution
The strains of HEV that exist today may have arisen from a shared ancestor virus 536 to 1344 years ago.[9] Another analysis has dated the origin of Hepatitis E to ~6000 years ago, with a suggestion that this was associated with domestication of pigs.[10] At some point, two clades may have diverged — an anthropotropic form and an enzootic form — which subsequently evolved into genotypes 1 and 2 and genotypes 3 and 4, respectively.[11]
Whereas genotype 2 remains less commonly detected than other genotypes, genetic evolutionary analyses suggest that genotypes 1, 3, and 4 have spread substantially during the past 100 years.[12]
See also
Notes
References
- 1 2 3 Purdy, Michael A.; et al. (June 2014). "New Classification Scheme for Hepeviridae" (PDF). International Committee on Taxonomy of Viruses (ICTV). Retrieved 1 May 2019.
The species Hepatitis E virus will be renamed Orthohepevirus A, and the species Avian hepatitis E virus will be renamed Orthohepevirus B.
- 1 2 3 4 "Hepeviridae - Hepeviridae - Positive-sense RNA Viruses - ICTV". www.ictv.global.
- ↑ Li P, Liu J, Li Y, Su J, Ma Z, Bramer WM, Cao W, de Man RA, Peppelenbosch MP, Pan Q (July 2020). "The global epidemiology of hepatitis E virus infection: A systematic review and meta-analysis". Liver International. 40 (7): 1516–1528. doi:10.1111/liv.14468. PMC 7384095. PMID 32281721.
- 1 2 Balayan MS, Andjaparidze AG, Savinskaya SS, et al. (1983). "Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal-oral route". Intervirology. 20 (1): 23–31. doi:10.1159/000149370. PMID 6409836.
- ↑ Reyes GR, Purdy MA, Kim JP, et al. (1990). "Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis". Science. 247 (4948): 1335–9. Bibcode:1990Sci...247.1335R. doi:10.1126/science.2107574. PMID 2107574.
- ↑ Schlauder, G. G. & Mushahwar, I. K. (2001) Genetic heterogeneity of hepatitis E virus. J Med Virol 65, 282–92
- ↑ Ahmad I, Holla RP, Jameel S (2011). "Molecular virology of hepatitis E virus". Virus Res. 161 (1): 47–58. doi:10.1016/j.virusres.2011.02.011. PMC 3130092. PMID 21345356.
- ↑ Osterman A, Stellberger T, Gebhardt A, Kurz M, Friedel CC, Uetz P, Nitschko H, Baiker A, Vizoso-Pinto MG (2015). "The Hepatitis E virus intraviral interactome". Sci Rep. 5: 13872. Bibcode:2015NatSR...513872O. doi:10.1038/srep13872. PMC 4604457. PMID 26463011.
- ↑ Khudyakov, Yury E.; Purdy, Michael A. (17 December 2010). "Evolutionary History and Population Dynamics of Hepatitis E Virus". PLOS ONE. 5 (12): e14376. Bibcode:2010PLoSO...514376P. doi:10.1371/journal.pone.0014376. ISSN 1932-6203. PMC 3006657. PMID 21203540.
- ↑ Baha, Sarra; Behloul, Nouredine; Liu, Zhenzhen; Wei, Wenjuan; Shi, Ruihua; Meng, Jihong (2019-10-29). "Comprehensive analysis of genetic and evolutionary features of the hepatitis E virus". BMC Genomics. 20 (1): 790. doi:10.1186/s12864-019-6100-8. ISSN 1471-2164. PMC 6820953. PMID 31664890.
- ↑ Mirazo S, Mir D, Bello G, Ramos N, Musto H, Arbiza J (2016). "New insights into the hepatitis E virus genotype 3 phylodynamics and evolutionary history". Infect Genet Evol. 43: 267–73. doi:10.1016/j.meegid.2016.06.003. PMID 27264728.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Izopet J, Abravanel F, Dalton H, Nassim RK (2014) Hepatitis E Virus Infection. Clin Micro Reviews 27 (1) 116–138