| Homoserine dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
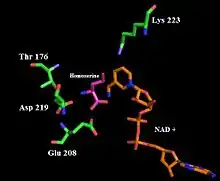
 Homoserine dehydrogenase complex with NAD+ analogue and L-homoserine. | |||||||||
| Identifiers | |||||||||
| Symbol | Homoserine_dh | ||||||||
| Pfam | PF00742 | ||||||||
| InterPro | IPR001342 | ||||||||
| PROSITE | PDOC00800 | ||||||||
| SCOP2 | 1ebu / SCOPe / SUPFAM | ||||||||
| |||||||||
| Homoserine dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Homoserine dehydrogenase homotetramer, Thiobacillus denitrificans | |||||||||
| Identifiers | |||||||||
| EC no. | 1.1.1.3 | ||||||||
| CAS no. | 9028-13-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
In enzymology, a homoserine dehydrogenase (EC 1.1.1.3) is an enzyme that catalyzes the chemical reaction
- L-homoserine + NAD(P)+ L-aspartate 4-semialdehyde + NAD(P)H + H+
The 2 substrates of this enzyme are L-homoserine and NAD+ (or NADP+), whereas its 3 products are L-aspartate 4-semialdehyde, NADH (or NADPH), and H+.
This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is L-homoserine:NAD(P)+ oxidoreductase. Other names in common use include HSDH, and HSD.
Homoserine dehydrogenase catalyses the third step in the aspartate pathway; the NAD(P)-dependent reduction of aspartate beta-semialdehyde into homoserine.[1][2] Homoserine is an intermediate in the biosynthesis of threonine, isoleucine, and methionine.[3]
Enzyme structure
The enzyme can be found in a monofunctional form, in some bacteria and yeast. Structural analysis of the yeast monofunctional enzyme indicates that the enzyme is a dimer composed of three distinct regions; an N-terminal nucleotide-binding domain, a short central dimerisation region, and a C-terminal catalytic domain.[4] The N-terminal domain forms a modified Rossmann fold, while the catalytic domain forms a novel alpha-beta mixed sheet.
The enzyme can also be found in a bifunctional form consisting of an N-terminal aspartokinase domain and a C-terminal homoserine dehydrogenase domain, as found in bacteria such as Escherichia coli and in plants.[5]
The bifunctional aspartokinase-homoserine dehydrogenase (AK-HSD) enzyme has a regulatory domain that consists of two subdomains with a common loop-alpha helix-loop-beta strand loop-beta strand motif. Each subdomain contains an ACT domain that allows for complex regulation of several different protein functions.[5] The AK-HSD gene codes for aspartate kinase, an intermediate domain (coding for the linker region between the two enzymes in the bifunctional form), and finally the coding sequence for homoserine dehydrogenase.[6][7]
As of late 2007, 4 structures have been solved for this class of enzymes, with PDB accession codes 1EBF, 1EBU, 1Q7G, and 1TVE.
Enzyme mechanism

Homoserine dehydrogenase catalyzes the reaction of aspartate-semialdehyde (ASA) to homoserine. The overall reaction reduces the C4 carboxylic acid functional group of ASA to a primary alcohol and oxidizes the C1 aldehyde to a carboxylic acid. Residues Glu 208 and Lys 117 are thought to be involved in the active catalytic site of the enzyme. Asp 214 and Lys 223 have been shown to be important for hydride transfer in the catalyzed reaction.[4]
Once the C4 carboxylic acid is reduced to an aldehyde and the C1 aldehyde is oxidized to a carboxylic acid, experiments suggest that Asp 219, Glu 208 and a water molecule bind ASA in the active site while Lys 223 donates a proton to the aspartate-semialdehyde C4 oxygen. Homoserine dehydrogenase has an NAD(P)H cofactor, which then donates a hydrogen to the same carbon, effectively reducing the aldehyde to an alcohol.[4] (Refer to figures 1 and 2).
However, the precise mechanism of complete homoserine dehydrogenase catalysis remains unknown.[4]
The homoserine dehydrogenase-catalyzed reaction has been postulated to proceed through a bi-bi kinetic mechanism, where the NAD(P)H cofactor binds the enzyme first and is the last to dissociate from the enzyme once the reaction is complete.[6][8] Additionally, while both NADH and NADPH are adequate cofactors for the reaction, NADH is preferred. The Km of the reaction is four-times smaller with NADH and the Kcat/Km is three-times greater, indicating a more efficient reaction.[9]
Homoserine dehydrogenase also exhibits multi-order kinetics at subsaturating levels of substrate. Additionally, the variable kinetics for homoserine dehydrogenase is an artifact of the faster dissociation of the amino acid substrate from the enzyme complex as compared to cofactor dissociation.[8][10]
Biological function
The aspartate metabolic pathway is involved in both storage of asparagine and in synthesis of aspartate-family amino acids.[11] Homoserine dehydrogenase catalyzes an intermediate step in this nitrogen and carbon storage and utilization pathway.[12] (Refer to figure 3).
In photosynthetic organisms, glutamine, glutamate, and aspartate accumulate during the day and are used to synthesize other amino acids. At night, aspartate is converted to asparagine for storage.[12] Additionally, the aspartate kinase-homoserine dehydrogenase gene is primarily expressed in actively growing, young plant tissues, particularly in the apical and lateral meristems.[13]
Mammals lack the enzymes involved in the aspartate metabolic pathway, including homoserine dehydrogenase. As lysine, threonine, methionine, and isoleucine are made in this pathway, they are considered essential amino acids for mammals.[6]
Biological regulation

Homoserine dehydrogenase and aspartate kinase are both subject to significant regulation (refer to figure 3). HSD is inhibited by downstream products of the aspartate metabolic pathway, mainly threonine. Threonine acts as a competitive inhibitor for both HSD and aspartate kinase.[14] In AK-HSD expressing organisms, one of the threonine binding sites is found in the linker region between AK and HSD, suggesting potential allosteric inhibition of both enzymes.[6]
However, some threonine-resistant HSD forms exist that require concentrations of threonine much greater than physiologically present for inhibition. These threonine-insensitive forms of HSD are used in genetically engineered plants to increase both threonine and methionine production for higher nutritional value.[6]
Homoserine dehydrogenase is also subject to transcriptional regulation. Its promoter sequence contains a cis-regulatory element TGACTC sequence, which is known to be involved in other amino acid biosynthetic pathways. The Opaque2 regulatory element has also been implicated in homoserine dehydrogenase regulation, but its effects are still not well defined.[7]
In plants, there is also environmental regulation of AK-HSD gene expression. Light exposure has been demonstrated to increase expression of the AK-HSD gene, presumably related to photosynthesis.[12][13]
Disease relevance
In humans, there has been a significant increase in disease from pathogenic fungi, so developing anti-fungal drugs is an important biochemical task.[15] As homoserine dehydrogenase is found mainly in plants, bacteria, and yeast, but not mammals, it is a strong target for antifungal drug development.[16] Recently, 5-hydroxy-4-oxonorvaline (HON) was discovered to target and inhibit HSD activity irreversibly. HON is structurally similar to aspartate semialdehyde, so it is postulated that it serves as a competitive inhibitor for HSD. Likewise, (S) 2-amino-4-oxo-5-hydroxypentanoic acid (RI-331), another amino acid analog, has also been shown to inhibit HSD.[16] Both of these compounds are effective against Cryptococcus neoformans and Cladosporium fulvum, among others.[17]
In addition to amino acid analogs, several phenolic compounds have been shown to inhibit HSD activity. Like HON and RI-331, these molecules are competitive inhibitors that bind to the enzyme active site. Specifically, the phenolic hydroxyl group interacts with the amino acid binding site.[15][18]
References
- ↑ Thomas D, Barbey R, Surdin-Kerjan Y (June 1993). "Evolutionary relationships between yeast and bacterial homoserine dehydrogenases". FEBS Lett. 323 (3): 289–93. doi:10.1016/0014-5793(93)81359-8. PMID 8500624. S2CID 23964791.
- ↑ Cami B, Clepet C, Patte JC (1993). "Evolutionary comparisons of three enzymes of the threonine biosynthetic pathway among several microbial species". Biochimie. 75 (6): 487–95. doi:10.1016/0300-9084(93)90115-9. PMID 8395899.
- ↑ Ferreira RR, Meinhardt LW, Azevedo RA (2006). "Lysine and threonine biosynthesis in sorghum seeds: characterisation of aspartate kinase and homoserine dehydrogenase isoenzymes". Ann. Appl. Biol. 149 (1): 77–86. doi:10.1111/j.1744-7348.2006.00074.x.
- 1 2 3 4 DeLaBarre B, Thompson PR, Wright GD, Berghuis AM (March 2000). "Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases". Nat. Struct. Biol. 7 (3): 238–44. doi:10.1038/73359. PMID 10700284. S2CID 26638309.
- 1 2 Paris S, Viemon C, Curien G, Dumas R (February 2003). "Mechanism of Control of Arabidopsis thaliana Aspartate Kinase-Homoserine Dehydrogenase by Threonine". J. Biol. Chem. 278 (7): 5361–5366. doi:10.1074/jbc.M207379200. PMID 12435751.
- 1 2 3 4 5 Schroeder AC, Zhu C, Yanamadala SR, Cahoon RE, Arkus KAJ, Wachsstock L, Bleeke J, Krishnan HB, Jez JM (January 2010). "Threonine-insensitive Homoserine Dehydrogenase from Soybean: Genomic Organization, Kinetic Mechanism, and in vivo Activity". J. Biol. Chem. 285 (2): 827–834. doi:10.1074/jbc.M109.068882. PMC 2801284. PMID 19897476.
- 1 2 Ghislain M, Frankard V, Vandenbossche D, Matthews BF, Jacobs M (March 1994). "Molecular analysis of the aspartate kinase-homoserine dehydrogenase gene from Arabidopsis thaliana". Plant Mol. Biol. 24 (6): 835–851. doi:10.1007/bf00014439. PMID 8204822. S2CID 6183867.
- 1 2 Wedler FC, Ley BW, Shames SL, Rembish SJ, Kushmaul DL (March 1992). "Preferred order random kinetic mechanism for homoserine dehydrogenase of Escherichia coli (Thr-sensitive) aspartokinase/homoserine dehydrogenase-I: Equilibrium Isotope Exchange Kinetics". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1119 (3): 247–249. doi:10.1016/0167-4838(92)90209-v. PMID 1547269.
- ↑ Jacques SL, Nieman C, Bareicha D, Broadhead G, Kinach R, Honek JF, Wright GD (January 2001). "Characterization of yeast homoserine dehydrogenase, an antifungal target: the invariant histidine 309 is important for enzyme integrity". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1544 (1–2): 28–41. doi:10.1016/S0167-4838(00)00203-X. PMID 11341914.
- ↑ Wedler FC, Ley BW (March 1993). "Kinetic and Regulatory Mechanisms for Escherichia coli Homoserine Dehydrogenase-I: Equilibrium Isotope Exchange Kinetics". J. Biol. Chem. 268 (1): 4880–4888. PMID 8444866.
- ↑ Azevedo RA (2002). "Analysis of the aspartic acid metabolic pathway using mutant genes". Amino Acids. 22 (3): 217–230. doi:10.1007/s007260200010. PMID 12083066. S2CID 23327489.
- 1 2 3 Zhu-Shimoni JX, Galili G (March 1998). "Expression of an Arabidopsis Aspartate Kinase/Homoserine Dehydrogenase Gene Is Metabolically Regulated by Photosynthesis-Related Signals but Not by Nitrogenous Compounds". Plant Physiol. 116 (3): 1023–1028. doi:10.1104/pp.116.3.1023. PMC 35071. PMID 9501134.
- 1 2 Zhu-Shimoni JX, Lev-Yadun S, Matthews B, Calili C (March 1997). "Expression of an Aspartate Kinase Homoserine Dehydrogenase Gene IS Subject to Specific Spatial and Temporal Regulation in Vegetative Tissues, Flowers, and Developing Seeds". Plant Physiol. 113 (3): 695–706. doi:10.1104/pp.113.3.695. PMC 158187. PMID 12223636.
- ↑ Park SD, Lee JY, Sim SY, Kim Y, Lee HS (July 2007). "Characteristics of methionine production by an engineered Corynebacterium glutamicum strain". Metab. Eng. 9 (4): 327–336. doi:10.1016/j.ymben.2007.05.001. PMID 17604670.
- 1 2 Bareich DC, Nazi I, Wright GD (October 2003). "Simultaneous In Vitro Assay of the First Four Enzymes in the Fungal Aspartate Pathway Identifies a New Class of Aspartate Kinase Inhibitor". Chem. Biol. 10 (10): 967–973. doi:10.1016/j.chembiol.2003.09.016. PMID 14583263.
- 1 2 Yamaki H, Yamaguchi M, Tsuruo T, Yamaguchi H (May 1992). "Mechanism of action of an antifungal antibiotich, RI-331, (S) 2-amino-4-oxo-5-hydroxypentanoic acid; kinetics of inactivation of homoserine dehydrogenase from Saccharomyces cerevisiae". J. Antibiot. (Tokyo). 45 (5): 750–755. doi:10.7164/antibiotics.45.750. PMID 1352515.
- ↑ Jacques SL, Mirza IA, Ejim L, Koteva K, Hughes DW, Green K, Kinach R, Honek JF, Lai HK, Berghuis AM, Wright GD (October 2003). "Enzyme-Assisted Suicide: Molecular Basis for the Antifungal Activity of 5-Hydroxy-4-Oxonorvaline by Potent Inhibition of Homoserine Dehydrogenase". Chem. Biol. 10 (10): 989–995. doi:10.1016/j.chembiol.2003.09.015. PMID 14583265.
- ↑ Ejim L, Mirza IA, Capone C, Nazi I, Jenkins S, Chee GL, Berghui AM, Wright GD (July 2004). "New phenolic inhibitors of yeast homoserine dehydrogenase". Bioorg. Med. Chem. 12 (14): 3825–3830. doi:10.1016/j.bmc.2004.05.009. PMID 15210149.
Further reading
- Black S, Wright NG (1955). "Homoserine dehydrogenase". J. Biol. Chem. 213 (1): 51–60. PMID 14353905.
- Starnes WL, Munk P, Maul SB, Cunningham GN, Cox DJ, Shive W (1972). "Threonine-sensitive aspartokinase-homoserine dehydrogenase complex, amino acid composition, molecular weight, and subunit composition of the complex". Biochemistry. 11 (5): 677–87. doi:10.1021/bi00755a003. PMID 4551091.
- Veron M, Falcoz-Kelly F, Cohen GN (1972). "The threonine-sensitive homoserine dehydrogenase and aspartokinase activities of Escherichia coli K12. The two catalytic activities are carried by two independent regions of the polypeptide chain". Eur. J. Biochem. 28 (4): 520–7. doi:10.1111/j.1432-1033.1972.tb01939.x. PMID 4562990.