 | |
 | |
| Names | |
|---|---|
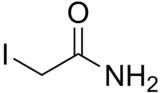
| Preferred IUPAC name
2-Iodoacetamide | |
| Other names
IAA[1] | |
| Identifiers | |
3D model (JSmol) |
|
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.005.119 |
| EC Number |
|
PubChem CID |
|
| RTECS number |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C2H4INO | |
| Molar mass | 184.964 g·mol−1 |
| Appearance | white crystals (yellow colouration indicates the presence of iodine) |
| Melting point | 94 °C (201 °F; 367 K) |
| Hazards | |
| NFPA 704 (fire diamond) | |
| Safety data sheet (SDS) | MSDS 1, MSDS 2 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
2-Iodoacetamide (IAA) is an alkylating agent used for peptide mapping purposes. Its actions are similar to those of iodoacetate. It is commonly used to bind covalently with the thiol group of cysteine so the protein cannot form disulfide bonds.[2][3] It is also used in ubiquitin studies as an inhibitor of deubiquitinase enzymes (DUBs) because it alkylates the cysteine residues at the DUB active site.
Peptidase inhibitor
Iodoacetamide is an irreversible inhibitor of all cysteine peptidases, with the mechanism of inhibition occurring from alkylation of the catalytic cysteine residue (see schematic). In comparison with its acid derivative, iodoacetate, iodoacetamide reacts substantially faster. This observation appears contradictory to standard chemical reactivity, however the presence of a favourable interaction between the positive imidazolium ion of the catalytic histidine and the negatively charged carboxyl-group of the iodoacetate is the reason for the increased relative activity of iodoacetamide.[4]

Protein mass spectrometry
It is commonly used during the sample preparation for de novo (peptide) sequencing with protein mass spectrometry, but recent critics suggest to avoid the use of it [5]
References
- ↑ Krüger, Ralf; Hung, Chien-Wen; Edelson-Averbukh, Marina; Lehmann, Wolf D. (2005). "Iodoacetamide-alkylated methionine can mimic neutral loss of phosphoric acid from phosphopeptides as exemplified by nano-electrospray ionization quadrupole time-of-flight parent ion scanning". Rapid Communications in Mass Spectrometry. Wiley. 19 (12): 1709–1716. doi:10.1002/rcm.1976. ISSN 0951-4198.
- ↑ Smythe CV (1936). "The reactions of Iodoacetate and of Iodoacetamide with various Sulfhydryl groups, with Urease, and with Yeast preparations". J. Biol. Chem. 114 (3): 601–12. doi:10.1016/S0021-9258(18)74789-3.
- ↑ Anson ML (1940). "The reactions of Iodine and Iodoacetamide with native Egg Albumin". J. Gen. Physiol. 23 (3): 321–31. doi:10.1085/jgp.23.3.321. PMC 2237930. PMID 19873158.
- ↑ Polgar, L (1979). "Deuterium isotope effects on papain acylation. Evidence for lack of general base catalysis and for enzyme-leaving group. interaction". Eur. J. Biochem. 98 (2): 369–374. doi:10.1111/j.1432-1033.1979.tb13196.x. PMID 488108.
- ↑ Müller (2017). "Systematic Evaluation of Protein Reduction and Alkylation Reveals Massive Unspecific Side Effects by Iodine-containing Reagents". Molecular & Cellular Proteomics. 16 (7): 1173–1187. doi:10.1074/mcp.M116.064048. PMC 5500753. PMID 28539326.
External links
- The MEROPS online database for peptidases and their inhibitors:
