| MSP (Major sperm protein) domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
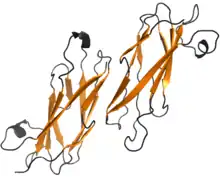 Structure of MSP dimer from A. suum. The β-sheets are shown in orange | |||||||||
| Identifiers | |||||||||
| Symbol | MSP_dom | ||||||||
| Pfam | PF00635 | ||||||||
| InterPro | IPR000535 | ||||||||
| PROSITE | PS50202 | ||||||||
| CATH | 1MSP | ||||||||
| |||||||||
| Major sperm protein | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Organism | |||||||
| Symbol | msp-19 | ||||||
| Entrez | 173981 | ||||||
| PDB | 1MSP | ||||||
| UniProt | P53017 | ||||||
| |||||||
Major sperm protein (MSP) is a nematode specific small protein of 126 amino acids with a molecular weight of 14 kDa. It is the key player in the motility machinery of nematodes that propels the crawling movement/motility of nematode sperm. It is the most abundant protein present in nematode sperm, comprising 15% of the total protein and more than 40% of the soluble protein. MSP is exclusively synthesized in spermatocytes of the nematodes.[1] The MSP has two main functions in the reproduction of the helminthes: i) as cytosolic component it is responsible for the crawling movement of the mature sperm (without flagellum), and ii) once released, it acts as hormone on the female germ cells, where it triggers oocyte maturation and stimulates the oviduct wall to contract to bring the oocytes into position for fertilization.[2] MSP has first been identified in Caenorhabditis elegans.
Structure
Molecular structures of MSP from Ascaris suum and Caenorhabditis elegans have been determined by X-ray crystallography[3] and NMR spectroscopy.[4][5] MSP molecules from these species share 83% sequence identity and their structures are highly similar.
MSP does not harbor any known conserved domain. It is made of a seven-stranded β sandwich, having opposing three-stranded and four-stranded β sheets. Hydrophobic side-chains from adjacent faces in the sandwich form the interior of the protein. The overall structure of MSP resembles an immunoglobulin fold (Ig fold). MSP can be classified as an s-type of this fold, because two of its strands are switching between separate β sheets, unlike in the conserved c-type of the Ig folds. The unique strand switches between the sheets result from two distinct kinks at cis-proline residues 13 and 57 in A. suum protein.[3]
MSP monomers form symmetric dimers. The interaction between MSP monomers in a dimer is very stable, with putative hydrophobic, hydrogen bond and salt bridge interactions. The residues involved in interface formation are between residue 13 and 29 in both A. suum MSP chains of the dimer.
MSP spontaneously polymerises both in vivo and in vitro from dimers into subfilaments, filaments, larger bundles and filament networks.[6]
MSP dimers are the smallest building blocks for these assemblies, none of which have overall polarity:
- subfilaments, formed from dimers, connected to a long helix. The dimer-dimer interface within the single subfilament is formed by residues 112-119 of two A. suum MSP chains, which produce an anti-parallel β-strand-β-strand pairing. The interaction is less hydrophobic and results mostly from formation of hydrogen bonds, typically for interfaces between reversibly interacting molecules.
- filaments, formed by two subfilaments coiling round one another. The MSP dimer-dimer interactions between two adjacent subfilaments in the filament are characterized by five interfaces, mostly between the residues 78-85 and 98-103. Amino acids 78-85 are part of a highly exposed surface loop connecting different β sheets and are divergent between C. elegans and A. suum. However, the loop consisting of 98-103 residues is highly conserved between all isoforms in both species of the nematode.
- fibers, macrofibers or bundles, produced by supercoiling of the filaments. A. suum MSP filaments frequently form rope-like structures called macrofibers. C. elegans MSP mostly form rafts in which a number of filaments are arranged parallel to one another.[4]
In contrast to actin, MSP lacks an ATP-binding site. However, it was noticed that ATP is required for MSP filament assembly at the surface of the plasma membrane. It was suggested that ATP activates either membrane-bound MSP filament end-tracking proteins or their soluble cofactors.[7]
Functions
Sperm motility
Nematode sperm move in an amoeboidal manner by extending a pseudopod.[8] Unlike the motility of actin-based cells, which is based on polar cytoskeletal elements such as actin monomers or tubulin dimers, nematode sperm locomotion is based on a pseudopod and a cytoskeleton built out of a meshwork of non-polar MSP filaments.[1][9][10] The two main differences between actin and MSP is that MSP does not bind ATP and the lack of polarity in MSP, thus disabling motility through motor proteins, such as myosin.
Locomotion in nematodes occurs by localized extension of the leading edge of the pseudopod, attachment of the cytoskeleton to the substrate, and retraction of the cell. Assembly of MSP filaments at the leading edge together with disassembly at the base of the pseudopod results in a treadmilling motion, which corresponds to the crawling locomotion of nematode sperm.[11]
Nematode sperm motility is based on a push-pull mechanism which requires two forces triggered by a pH gradient along the pseudopod: one protrusive force and another traction force. The protrusive force is located at the leading edge and pushes against the cell membrane. This force is generated by polymerization of the MSP filaments. MSP filaments are assembled in the cytoplasm near the leading edge of the pseudopod out of MSP dimers resulting in extensions. These extensions allow the interaction of the filament complexes with surrounding complexes, thus resulting in an interconnected uniform cytoskeleton and leading to the crawling movement of the cytoskeleton. MSP filament assembly is triggered by external factors, such as changes in pH,[12] the integral membrane phosphoprotein (MPOP), and MSP domain proteins (MDPs).[11][13][14]
A 48 kDa integral membrane phosphoprotein, the major sperm protein polymerization organization protein (MPOP), is the starting point of the pseudopod and is required for the localized membrane-associated polymerization of MSP.[15] This protein is distributed in vesicles throughout the pseudopodal membrane. Tyrosine kinases, which are pH sensitive, phosphorylate the tyrosine residues of MPOP localized at the tip of the pseudopod, thus resulting in the polymerization of the MSP filaments.[16] In Ascaris suum, two MSP-fiber proteins (MFP), MFP1 and 2, with opposite effect on polymerization have been identified. MFP1 inhibits and MFP2 stimulates the MSP assembly.[17] Changes in pH both controls and activates the MSP polymerization throughout spermatogenesis by a pH gradient within the pseudopod of the spermatozoon: assembly occurs at the leading edge where the pH is high, and disassembly of the filaments occurs at the base where the pH is lowered.[11] Degradation of the MSP filaments results in a traction force at the base of the pseudopod, which in turn pulls the cytoskeleton forward. The combination of these two forces is the motive force that allows sperm motility. Attachment of the cytoskeleton to the substratum is required to generate a directional movement.[18][19]
Affecting female germ cells
MSP affects oocytes on two levels:
- MSP regulates oocyte maturation. In C. elegans, oocytes arrest their meiotic cycle at metaphase of meiosis I where it is only resumed in presence of sperms. MSP was identified as the molecular factor triggering oocyte meiotic maturation. It is secreted by the sperms through a vesicular budding mechanism and forms an extracellular concentration gradient. MSP binds to VAB-1 which is an Eph receptor protein-tyrosine kinase on oocytes. In absence of MSP, the VAB-1 Eph receptor inhibits meiotic maturation of oocytes through interaction with inhibitors DAB-1/Disabled and RAN-1. MSP binding prevents this inhibition and results in activation of MAPK pathway.[2][20][21][22]
- MSP also stimulates gonadal sheath cell contraction which is a myoepithelial sheath surrounding proximal oocytes. It increases the contraction rate from 10-13 to around 19 contractions per minute. The importance of these contractions is promoting ovulation by inducing the enveloping of the oocyte by the spermatheca.[23]
Homologues
MSP genes have been identified across widely diverged nematode species. They all have more than 60% sequence identity.[1]
Proteins with limited sequence similarity were identified in species from plants to mammals. One of the homologues is VAP33 from Aplysia californica.[24] VAP33 is a protein required for neurotransmitter release, which binds to the v-SNARE synaptobrevin/VAMP, associated with vesicle fusion.
Despite only 11% of sequence similarity, MSP and the N-terminus of the bacterial P-pilus associated chaperonin PapD share a high structural and topological homology in their β sheet regions. Both MSP and PapD can be classified to the s-type immunoglobulin fold proteins, characterized by the above-mentioned unique strand switching.[3]
References
- 1 2 3 Scott AL (November 1996). "Nematode sperm". Parasitol. Today (Regul. Ed.). 12 (11): 425–30. doi:10.1016/0169-4758(96)10063-6. PMID 15275275.
- 1 2 Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D (March 2001). "A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation". Science. 291 (5511): 2144–7. doi:10.1126/science.1057586. PMID 11251118.
- 1 2 3 Bullock TL, Roberts TM, Stewart M (October 1996). "2.5 Å resolution crystal structure of the motile major sperm protein (MSP) of Ascaris suum". J. Mol. Biol. 263 (2): 284–96. doi:10.1006/jmbi.1996.0575. PMID 8913307.
- 1 2 Haaf A, LeClaire L, Roberts G, Kent HM, Roberts TM, Stewart M, Neuhaus D (December 1998). "Solution structure of the motile major sperm protein (MSP) of Ascaris suum - evidence for two manganese binding sites and the possible role of divalent cations in filament formation". J. Mol. Biol. 284 (5): 1611–24. doi:10.1006/jmbi.1998.2291. PMID 9878374.
- ↑ Baker AM, Roberts TM, Stewart M (May 2002). "2.6 A resolution crystal structure of helices of the motile major sperm protein (MSP) of Caenorhabditis elegans". J. Mol. Biol. 319 (2): 491–9. doi:10.1016/S0022-2836(02)00294-2. PMID 12051923.
- ↑ King KL, Stewart M, Roberts TM, Seavy M (April 1992). "Structure and macromolecular assembly of two isoforms of the major sperm protein (MSP) from the amoeboid sperm of the nematode, Ascaris suum". J. Cell Sci. 101. ( Pt 4): 847–57. PMID 1527183.
- ↑ Dickinson RB, Purich DL (January 2007). "Nematode sperm motility: nonpolar filament polymerization mediated by end-tracking motors". Biophys. J. 92 (2): 622–31. doi:10.1529/biophysj.106.090472. PMC 1751402. PMID 17056726.
- ↑ Roberts TM, Stewart M (February 1995). "Nematode sperm locomotion". Curr. Opin. Cell Biol. 7 (1): 13–7. doi:10.1016/0955-0674(95)80039-5. PMID 7755984.
- ↑ Roberts TM, Stewart M (April 2000). "Acting like actin. The dynamics of the nematode major sperm protein (msp) cytoskeleton indicate a push-pull mechanism for amoeboid cell motility". J. Cell Biol. 149 (1): 7–12. doi:10.1083/jcb.149.1.7. PMC 2175093. PMID 10747081.
- ↑ Roberts TM, Salmon ED, Stewart M (January 1998). "Hydrostatic pressure shows that lamellipodial motility in Ascaris sperm requires membrane-associated major sperm protein filament nucleation and elongation". J. Cell Biol. 140 (2): 367–75. doi:10.1083/jcb.140.2.367. PMC 2132582. PMID 9442112.
- 1 2 3 Bottino D, Mogilner A, Roberts T, Stewart M, Oster G (January 2002). "How nematode sperm crawl". J. Cell Sci. 115 (Pt 2): 367–84. PMID 11839788.
- ↑ Italiano JE, Stewart M, Roberts TM (September 1999). "Localized depolymerization of the major sperm protein cytoskeleton correlates with the forward movement of the cell body in the amoeboid movement of nematode sperm". J. Cell Biol. 146 (5): 1087–96. doi:10.1083/jcb.146.5.1087. PMC 2169480. PMID 10477761.
- ↑ Wolgemuth CW, Miao L, Vanderlinde O, Roberts T, Oster G (April 2005). "MSP dynamics drives nematode sperm locomotion". Biophys. J. 88 (4): 2462–71. doi:10.1529/biophysj.104.054270. PMC 1305345. PMID 15665134.
- ↑ Tarr DE, Scott AL (May 2005). "MSP domain proteins". Trends Parasitol. 21 (5): 224–31. doi:10.1016/j.pt.2005.03.009. PMID 15837611.
- ↑ LeClaire LL, Stewart M, Roberts TM (July 2003). "A 48 kDa integral membrane phosphoprotein orchestrates the cytoskeletal dynamics that generate amoeboid cell motility in Ascaris sperm". J. Cell Sci. 116 (Pt 13): 2655–63. doi:10.1242/jcs.00469. PMID 12746486.
- ↑ Yi K, Buttery SM, Stewart M, Roberts TM (May 2007). "A Ser/Thr kinase required for membrane-associated assembly of the major sperm protein motility apparatus in the amoeboid sperm of Ascaris". Mol. Biol. Cell. 18 (5): 1816–25. doi:10.1091/mbc.E06-08-0741. PMC 1855020. PMID 17344482.
- ↑ Buttery SM, Ekman GC, Seavy M, Stewart M, Roberts TM (December 2003). "Dissection of the Ascaris sperm motility machinery identifies key proteins involved in major sperm protein-based amoeboid locomotion". Mol. Biol. Cell. 14 (12): 5082–8. doi:10.1091/mbc.E03-04-0246. PMC 284809. PMID 14565983.
- ↑ Miao L, Vanderlinde O, Stewart M, Roberts TM (November 2003). "Retraction in amoeboid cell motility powered by cytoskeletal dynamics". Science. 302 (5649): 1405–7. doi:10.1126/science.1089129. PMID 14631043.
- ↑ Roberts TM (March 2005). "Major sperm protein". Curr. Biol. 15 (5): R153. doi:10.1016/j.cub.2005.02.036. PMID 15753021.
- ↑ Govindan JA, Nadarajan S, Kim S, Starich TA, Greenstein D (July 2009). "Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans". Development. 136 (13): 2211–21. doi:10.1242/dev.034595. PMC 2729340. PMID 19502483.
- ↑ Greenstein D (2005). "Control of oocyte meiotic maturation and fertilization". WormBook: 1–12. doi:10.1895/wormbook.1.53.1. PMC 4781623. PMID 18050412.
- ↑ Cheng H, Govindan JA, Greenstein D (May 2008). "Regulated trafficking of the MSP/Eph receptor during oocyte meiotic maturation in C. elegans". Curr. Biol. 18 (10): 705–14. doi:10.1016/j.cub.2008.04.043. PMC 2613949. PMID 18472420.
- ↑ Kuwabara PE (January 2003). "The multifaceted C. elegans major sperm protein: an ephrin signaling antagonist in oocyte maturation". Genes Dev. 17 (2): 155–61. doi:10.1101/gad.1061103. PMID 12533505.
- ↑ Skehel PA, Martin KC, Kandel ER, Bartsch D (September 1995). "A VAMP-binding protein from Aplysia required for neurotransmitter release". Science. 269 (5230): 1580–3. doi:10.1126/science.7667638. PMID 7667638.