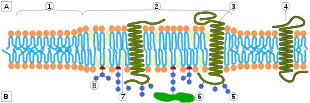
The plasma membranes of cells contain combinations of glycosphingolipids, cholesterol and protein receptors organised in glycolipoprotein lipid microdomains termed lipid rafts.[1][2][3] Their existence in cellular membranes remains somewhat controversial. It has been proposed that they are specialized membrane microdomains which compartmentalize cellular processes by serving as organising centers for the assembly of signaling molecules, allowing a closer interaction of protein receptors and their effectors to promote kinetically favorable interactions necessary for the signal transduction.[4] Lipid rafts influence membrane fluidity and membrane protein trafficking, thereby regulating neurotransmission and receptor trafficking.[3][5] Lipid rafts are more ordered and tightly packed than the surrounding bilayer, but float freely within the membrane bilayer.[6] Although more common in the cell membrane, lipid rafts have also been reported in other parts of the cell, such as the Golgi apparatus and lysosomes.
Properties
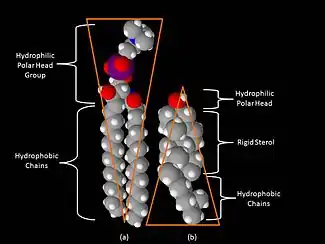
One key difference between lipid rafts and the plasma membranes from which they are derived is lipid composition. Research has shown that lipid rafts contain 3 to 5-fold the amount of cholesterol found in the surrounding bilayer.[7] Also, lipid rafts are enriched in sphingolipids such as sphingomyelin, which is typically elevated by 50% compared to the plasma membrane. To offset the elevated sphingolipid levels, phosphatidylcholine levels are decreased which results in similar choline-containing lipid levels between the rafts and the surrounding plasma membrane. Cholesterol interacts preferentially, although not exclusively, with sphingolipids due to their structure and the saturation of the hydrocarbon chains. Although not all of the phospholipids within the raft are fully saturated, the hydrophobic chains of the lipids contained in the rafts are more saturated and tightly packed than the surrounding bilayer.[5] Cholesterol is the dynamic "glue" that holds the raft together.[3] Due to the rigid nature of the sterol group, cholesterol partitions preferentially into the lipid rafts where acyl chains of the lipids tend to be more rigid and in a less fluid state.[5] One important property of membrane lipids is their amphipathic character. Amphipathic lipids have a polar, hydrophilic head group and a non-polar, hydrophobic region.[8] The figure to the right shows the inverted cone-like shape of sphingomyelin and the cone-like shape of cholesterol based on the area of space occupied by the hydrophobic and hydrophilic regions. Cholesterol can pack in between the lipids in rafts, serving as a molecular spacer and filling any voids between associated sphingolipids.[8]
Rietveld & Simons related lipid rafts in model membranes to the immiscibility of ordered (Lo phase) and disordered (Ld or Lα phase) liquid phases.[9] The cause of this immiscibility is uncertain, but the immiscibility is thought to minimize the free energy between the two phases. Studies have shown there is a difference in thickness of the lipid rafts and the surrounding membrane which results in hydrophobic mismatch at the boundary between the two phases. This phase height mismatch has been shown to increase line tension which may lead to the formation of larger and more circular raft platforms to minimize the energetic cost of maintaining the rafts as a separate phase. Other spontaneous events, such as curvature of the membrane and fusing of small rafts into larger rafts, can also minimize line tension.[5]
By one early definition of lipid rafts, lipid rafts differ from the rest of the plasma membrane. In fact, researchers[10] have hypothesized that the lipid rafts can be extracted from a plasma membrane. The extraction would take advantage of lipid raft resistance to non-ionic detergents, such as Triton X-100 or Brij-98 at low temperatures (e.g., 4 °C). When such a detergent is added to cells, the fluid membrane will dissolve while the lipid rafts may remain intact and could be extracted.
Because of their composition and detergent resistance, lipid rafts are also called detergent-insoluble glycolipid-enriched complexes (GEMs) or DIGs[11] or Detergent Resistant Membranes (DRMs). However the validity of the detergent resistance methodology of membranes has recently been called into question due to ambiguities in the lipids and proteins recovered and the observation that they can also cause solid areas to form where there were none previously.[12]
Function
Mediation of substrate presentation. Lipid rafts localize palmitoylated proteins away from the disordered region of the plasma membrane.[13] Disruption of palmitate mediated localization then allows for exposure of a protein to its binding partner or substrate in the disordered region, an activation mechanism termed substrate presentation. For example, a protein is often palmitoylated and binds phosphatidylinositol 4,5-biphosphate (PIP2). PIP2 is polyunsaturated and does not reside in lipid rafts. When the levels of PIP2 increase in the plasma membrane, the protein trafficks to PIP2 clusters where it can be activated directly by PIP2 (or another molecule that associates with PIP2).[14][15]
It is probable that other functions exist.
History
Until 1982, it was widely accepted that phospholipids and membrane proteins were randomly distributed in cell membranes, according to the Singer-Nicolson fluid mosaic model, published in 1972.[5][16] However, membrane microdomains were postulated in the 1970s using biophysical approaches by Stier & Sackmann[17] and Klausner & Karnovsky.[18] These microdomains were attributed to the physical properties and organization of lipid mixtures by Stier & Sackmann and Israelachvili et al.[19] In 1974, the effects of temperature on membrane behavior had led to the proposal of "clusters of lipids" in membranes and by 1975, data suggested that these clusters could be "quasicrystalline" regions within the more freely dispersed liquid crystalline lipid molecule. In 1978, X-Ray diffraction studies led to further development of the "cluster" idea defining the microdomains as "lipids in a more ordered state". Karnovsky and co-workers formalized the concept of lipid domains in membranes in 1982. Karnovsky's studies showed heterogeneity in the lifetime decay of 1,6-diphenyl-1,3,5-hexatriene, which indicated that there were multiple phases in the lipid environment of the membrane.[5] One type of microdomain is constituted by cholesterol and sphingolipids. They form because of the segregation of these lipids into a separate phase, demonstrated by Biltonen and Thompson and their coworkers.[20] These microdomains (‘rafts’) were shown to exist also in cell membranes.[21] Later, Kai Simons at the European Molecular Biology Laboratory (EMBL) in Germany and Gerrit van Meer from the University of Utrecht, Netherlands refocused interest on these membrane microdomains, enriched with lipids and cholesterol, glycolipids, and sphingolipids, present in cell membranes.[22] Subsequently, they called these microdomains, lipid "rafts". The original concept of rafts was used as an explanation for the transport of cholesterol from the trans Golgi network to the plasma membrane. The idea was more formally developed in 1997 by Simons and Ikonen.[23] At the 2006 Keystone Symposium of Lipid Rafts and Cell Function, lipid rafts were defined as "small (10-200nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein interactions" In recent years, lipid raft studies have tried to address many of the key issues that cause controversy in this field, including the size and lifetime of rafts.
Other questions yet to be answered include:
- What are the effects of membrane protein levels?
- What is the physiological function of lipid rafts?
- What effect does flux of membrane lipids have on raft formation?
- What effect do diet and drugs have on lipid rafts?
- What effect do proteins located at raft boundaries have on lipid rafts?[5]
Common types
Two types of lipid rafts have been proposed: planar lipid rafts (also referred to as non-caveolar, or glycolipid, rafts) and caveolae. Planar rafts are defined as being continuous with the plane of the plasma membrane (not invaginated) and by their lack of distinguishing morphological features. Caveolae, on the other hand, are flask shaped invaginations of the plasma membrane that contain caveolin proteins and are the most readily-observed structures in lipid rafts. Caveolins are widely expressed in the brain, micro-vessels of the nervous system, endothelial cells, astrocytes, oligodendrocytes, Schwann cells, dorsal root ganglia and hippocampal neurons. Planar rafts contain flotillin proteins and are found in neurons where caveolae are absent. Both types have similar lipid composition (enriched in cholesterol and sphingolipids). Flotillin and caveolins can recruit signaling molecules into lipid rafts, thus playing an important role in neurotransmitter signal transduction. It has been proposed that these microdomains spatially organize signaling molecules to promote kinetically favorable interactions which are necessary for signal transduction. Conversely, these microdomains can also separate signaling molecules, inhibiting interactions and dampening signaling responses.[24]
Role in signal transduction
The specificity and fidelity of signal transduction are essential for cells to respond efficiently to changes in their environment. This is achieved in part by the differential localization of proteins that participate in signalling pathways. In the plasma membrane, one approach of compartmentalization utilizes lipid rafts.[25]
One reasonable way to consider lipid rafts is that small rafts can form concentrating platforms after ligand binding activation for individual receptors.[26] Lipid rafts have been found by researchers to be involved in many signal transduction processes, such as Immunoglobulin E signalling, T cell antigen receptor signalling, B cell antigen receptor signalling, EGF receptor signalling, insulin receptor signalling and so on. In order to illustrate these principles, detailed examples of signalling pathways that involve lipid rafts are described below.
Epidermal growth factor signaling
Epidermal growth factor (EGF) binds to EGF receptor, also known as HER-1 or ErbB1, to initiate transmembrane signaling. Lipid rafts have been suggested to play a bipartite role in this process. Certain aspects of lipid rafts inhibit EGF receptor function:
- the ganglioside component of lipid rafts was shown to inhibit receptor activation[27][28]
- the membrane dipole potential, which was shown to be higher in lipid rafts than in the rest of the membrane,[29] was demonstrated to inhibit EGF binding to its receptor[30]
- EGF binding was shown to be inhibited by non-caveolar lipid rafts due to a decrease in the number of receptors available for ligand binding[31]
- EGF[32] and ErbB2 (HER-2)[30] were shown to migrate out of lipid rafts or caveolae during or after activation
- disruption of lipid rafts was shown to induce ligand-independent activation of EGF receptor[33]
At the same time lipid rafts seem to be necessary for or potentiate transmembrane signaling:
- sequestration of ErbB2 from lipid rafts have been shown to inhibit EGF-induced signaling[34]
- the membrane dipole potential, which is higher in lipid rafts than in the rest of the membrane,[29] potentiates EGF-induced signaling[30]
- EGF was shown to bring about coalescence of individual lipid rafts,[35] similar to what has been suggested to play a role in the activation of the T-cell receptor[36]
- localization of EGF receptor to lipid rafts induces resistance to tyrosine kinase inhibitors[37]
Immunoglobulin E signaling

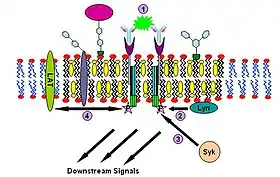
Immunoglobulin E (IgE) signaling is the first convincingly demonstrated lipid rafts involving signaling process.[38][39][40] Evidence for this fact includes decreased solubility of Fc-epsilon receptors (FcεR) in Triton X-100 from steady state to crosslinking state,[38] formation of patches large enough to be visualized by fluorescence microscopy from gangliosides and GPI-anchored proteins,[41][42] abolition of IgE signaling by surface cholesterol depletion with methyl-β-cyclodextrin[43] and so on. This signaling pathway can be described as follows: IgE first binds to Fc-epsilon receptors (FcεR) residing in the plasma membrane of mast cells and basophils through its Fc segment. FcεR is a tetramer consist of one α, one β and two γ chains.[40] It is monomeric and binds one IgE molecule. The α chain binds IgE and the other three chains contain immune receptor tyrosine-based activation motifs (ITAM). Then oligomeric antigens bind to receptor-bound IgE to crosslink two or more of these receptors. This crosslinking then recruits doubly acylated non-receptor Src-like tyrosine kinase Lyn to phosphorylate ITAMs. After that, Syk family tyrosine kinases bind these phosphotyrosine residues of ITAMs to initiate the signaling cascade.[38][39] Syk can, in turn, activate other proteins such as LAT. Through crosslinking, LAT can recruit other proteins into the raft and further amplify the signal.[44]
T-cell antigen receptor signaling

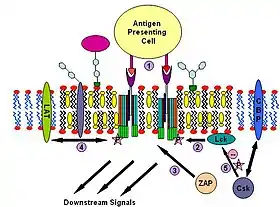
T cell antigen receptor (TCR) is a molecule found on the surface of T lymphocytes (T cells). It is composed of αβ-heterodimers, CD3 (γδε) complex and ξ-homodimer. The α- and β- subunits contains extracellular binding sites for peptides that are presented by the major histocompatibility complex (MHC) class I and class II proteins on the surface of antigen presenting cells (APCs). The CD3 and ξ- subunits contain cytoplasmic ITAM motifs. During the signaling process, MHCs binding to TCRs brings two or more receptors together. This crosslinking, similar to IgE signaling, then recruit doubly acylated non-receptor Src-like tyrosine kinases to phosphorylate ITAM tyrosine residues. In addition to recruiting Lyn, TCR signaling also recruits Fyn.[25][45] Following this procedure, ZAP-70 (which is also different with IgE signalling) binds to phosphorylated ITAMs, which leads to its own activation and LAT activation. LAT activation is the source of signal amplification. Another difference between IgE and T cell antigen receptor signalling is that Lck activation by TCR could result in more severe raft clustering[46][47] thus more signal amplification. One possible mechanism of down-regulating this signaling involves the binding of cytosolic kinase Csk to the raft associated protein CBP. Csk may then suppress the Src-family kinases through phosphorylation.[48]
B-cell antigen receptor signaling
B cell antigen receptor (BCR) is a complex between a membrane bound Ig (mIg) molecule and a disulfide-linked Igα- Igβ heterodimer of two polypeptides.[49] Igα and Igβ each contains an amino acid motif, called ITAM, whose sequence is D/ExxYxxL/Ix7YxxL/I.
The process of B cell antigen receptor signalling is similar to Immunoglobulin E signalling and T-cell antigen receptor signalling. It is commonly believed that other than BCR, lipid rafts play an important role in many of the cell surface events involved in B cell activation. Their functions include signaling by BCR, modulation of that signaling by co-receptors, signaling by CD40, endocytosis of antigen bound to the BCR and its routing to late endosomes to facilitate loading of antigen-derived peptides onto class II MHC molecules, routing of those peptide/MHC-II complexes to the cell surface, and their participation in antigen presentation to T cells.[49]
As platforms for virus entry
Viruses, as obligate intracellular parasites, have to involve specific interaction of virus and cellular receptor expressed at the plasma membrane in order to enter cells. Accumulated evidence supports that viruses enter cells via penetration of specific membrane microdomains, including lipid rafts.
Nonenveloped virus
The best studied models of lipid rafts-related nonenveloped viral entry are simian virus 40 (SV40, Papovaviridae) and echovirus type 1 (EV1, Picornaviridae).[50][51]
SV40 utilizes two different receptors to bind onto cell surface: ganglioside GM1 located in lipid rafts and major histocompatibility (MHC) class I molecule.[50][51] Binding of SV40 with MHC class I molecules triggers receptor clustering and redistribution. SV40 may recruit more caveolae from the cytoplasm or even new caveolae formed at the site of entry.[51] A cascade of virus-induced signaling events triggered by attachment results in caveolae-mediated endocytosis in about 20 min.[51] In some cell types the virus can enter the caveosomes directly from lipid rafts in non-coated vesicles.[51][52]
EV1 uses α2β1-integrin as cellular receptor.[50] Multiple integrin heterodimers can bind to the adjacent sites of the virus capsid.[51] Similar to SV40, attachment and binding with cells triggers clustering and relocation of integrin molecules from lipid rafts to the caveolae-like structures.[51] Depletion of cholesterol in lipid rafts inhibits EV1 infection.[50]
There are also viruses that use the non-caveolar raft-mediated endocytosis, such as Echovirus 11 (EV11, picornavirus). However, detailed mechanisms still need to be further characterized.[51]
Enveloped virus
Influenza viruses bind to the cellular receptor sialic acid, which links to glycoconjugate on the cell surface, to initiate endocytosis. After transportation into late endosomes, low-pH-dependent conformation changes of HA induces fusion, and viral ribonucleoprotein complexes (RNP) are released by proton influx of viral ion channel M2 proteins that requires binding with cholesterol. Semliki Forest virus (SFV) and Sindbis virus (SIN) require cholesterol and sphingolipids in target membrane lipid rafts for envelope glycoprotein-mediated membrane fusion and entry.[53] Human T-lymphotropic virus Type I (HTLV-1) enter cells via glucose transporter 1 (GLUT-1). Ebola virus and Marburg virus use folate receptor-α (FRα), which is a GPI-anchored protein, as a cellular receptor. Hepatitis B virus recognizes human complement receptor type 2 (CR2, or known as CD21). Human herpesvirus 6 (HHV-6) binds to human CD46 on host cell surface. All these viral receptors are located in lipid rafts or would be relocated into lipid rafts after infection.
Human Immunodeficiency virus (HIV), as a sexually-transmitted animal virus, must first penetrate a barrier of epithelial cells, who don't express CD4 and chemokine receptors, to establish a productive infection. An alternative receptor for HIV-1 envelope glycoprotein on epithelial cells is glycosphingolipid galactosyl-ceramide (GalCer), which enriches at lipid raft.[54][55]
SARS-Cov-2
The SARS-CoV-2 virus that causes COVID-19 was shown to enter through endocytosis using lipid rafts.[56] The omicron variant predominantly enters through endocytosis, presumably through lipid rafts.[57] Hydroxychloroquine blocks the entry of SARS-CoV-2 by blocking ACE2 association with enodocytic lipids.[58]
Visualization
One of the primary reasons for the controversy over lipid rafts has stemmed from the challenges of studying lipid rafts in living cells, which are not in thermodynamic equilibrium.[24] Lipid rafts are small microdomains ranging from 10 to 200 nm in size.[5] Due to their size being below the classical diffraction limit of a light microscope, lipid rafts have proved difficult to visualize directly. Currently synthetic membranes are studied; however, there are many drawbacks to using these membranes. First, synthetic membranes have a lower concentration of proteins compared to biomembranes. Also, it is difficult to model membrane-cytoskeletal interactions which are present in biomembranes. Other pitfalls include lack of natural asymmetry and inability to study the membranes in non-equilibrium conditions.[5][59] Despite this, fluorescence microscopy is used extensively in the field. For example, fluorophores conjugated to cholera-toxin B-subunit, which binds to the raft constituent ganglioside GM1 is used extensively. Also used are lipophilic membrane dyes which either partition between rafts and the bulk membrane, or change their fluorescent properties in response to membrane phase. Laurdan is one of the prime examples of such a dye. Rafts may also be labeled by genetic expression of fluorescent fusion proteins such as Lck-GFP.
Manipulation of cholesterol is one of the most widely used techniques for studying lipid rafts. Sequestration (using filipin, nystatin or amphotericin), depletion and removal (using methyl-B-cyclodextrin) and inhibition of cholesterol synthesis (using HMG-CoA reductase inhibitors) are ways cholesterol are manipulated in lipid raft studies. These studies allow for the observations of effects on neurotransmitter signaling upon reduction of cholesterol levels.[24]
Sharma and colleagues used combination of high resolution imaging and mathematical modeling to provide the view that raft proteins are organized into high density nanoclusters with radii ranging over 5–20 nm. Using measurements of fluorescence resonance energy transfer between the same probes (homo-FRET or fluorescence anisotropy), Sharma and colleagues reported that a fraction (20–40%) of GPI-anchored proteins are organized into high density clusters of 4–5 nm radius, each consisting of a few molecules and different GPI-anchored proteins.[60] To combat the problems of small size and dynamic nature, single particle and molecule tracking using cooled, sensitive CCD cameras and total internal reflection (TIRF) microscopy is coming to prominence. This allows information of the diffusivity of particles in the membrane to be extracted as well as revealing membrane corrals, barriers and sites of confinement.[61]
Other optical techniques are also used: Fluorescence Correlation and Cross-Correlation Spectroscopy (FCS/FCCS) can be used to gain information of fluorophore mobility in the membrane, Fluorescence Resonance Energy Transfer (FRET) can detect when fluorophores are in close proximity and optical tweezer techniques can give information on membrane viscosity.[24]
Not only optical techniques, but also scanning probe techniques like atomic force microscopy (AFM) or Scanning Ion Conductance Microscopy (SICM) can be used to detect the topological and mechanical properties of synthetic lipids[62] or native cell membranes[63] isolated by cell unroofing.
Also used are dual polarisation interferometry, Nuclear Magnetic Resonance (NMR) although fluorescence microscopy remains the dominant technique. In the future it is hoped that super-resolution microscopy such as Stimulated Emission Depletion (STED)[64] or various forms of structured illumination microscopy may overcome the problems imposed by the diffraction limit.
Other techniques used in the analysis of lipid rafts include ELISA, western blotting, and FACS.[65][66][1]
Controversy
The role of rafts in cellular signaling, trafficking, and structure has yet to be determined despite many experiments involving several different methods, and their very existence is controversial despite all the above.[67]
Arguments against the existence of lipid rafts include the following:
- First, a line tension should exist between the Lα and Lo phases. This line has been seen in model membranes, but has not been readily observed in cell systems.
- Second, there is no consensus on lipid raft size, which has been reported anywhere between 1 and 1,000 nanometers.
- Third, the time scale of lipid raft existence is unknown. If lipid rafts exist, they may only occur on a time scale that is irrelevant to biological processes.
- Fourth, the entire membrane may exist in the Lo phase.
A first rebuttal to this point suggests that the Lo phase of the rafts is more tightly packed due to the intermolecular hydrogen bonding exhibited between sphingolipids and cholesterol that is not seen elsewhere.[68]
A second argument questions the effectiveness of the experimental design when disrupting lipid rafts. Pike and Miller discuss potential pitfalls of using cholesterol depletion to determine lipid raft function.[69] They noted that most researchers were using acute methods of cholesterol depletion, which disrupt the rafts, but also disrupt another lipid known as PI(4,5)P2. PI(4,5)P2 plays a large role in regulating the cell's cytoskeleton,[70] and disrupting PI(4,5)P2 causes many of the same results as this type of cholesterol depletion, including lateral diffusion of the proteins in the membrane.[71] Because the methods disrupt both rafts and PI(4,5)P2, Kwik et al. concluded that loss of a particular cellular function after cholesterol depletion cannot necessarily be attributed solely to lipid raft disruption, as other processes independent of rafts may also be affected. Finally, while lipid rafts are believed to be connected in some way to proteins, Edidin argues that proteins attract the lipids in the raft by interactions of proteins with the acyl chains on the lipids, and not the other way around.[72]
References
- 1 2 Thomas, Sunil; Preda-Pais, Anca; Casares, Sofia; Brumeanu, Teodor-D (2004). "Analysis of lipid rafts in T cells". Molecular Immunology. 41 (4): 399–409. doi:10.1016/j.molimm.2004.03.022. PMID 15163537.
- ↑ Thomas, Sunil; Kumar S., Rajeev; Brumeanu, Teodor−D. (2004). "Role of lipid rafts in T cells". Archivum Immunologiae et Therapiae Experimentalis. 52 (4): 215–24. PMID 15467486.
- 1 2 3 Korade, Zeljka; Kenworthy, Anne K. (2008). "Lipid rafts, cholesterol, and the brain". Neuropharmacology. 55 (8): 1265–73. doi:10.1016/j.neuropharm.2008.02.019. PMC 2638588. PMID 18402986.
- ↑ Alves, Anna Carolina Schneider; Dias, Reinaldo Antonio; Kagami, Luciano Porto; Neves, Gustavo Machado das; Torres, Fernando Cidade; Eifler-Lima, Vera Lucia; Carvalho, Ivone; Kawano*, Carolina de Miranda Silva and Daniel Fabio (2018-05-31). "Beyond the". Current Medicinal Chemistry. 25 (18): 2082–2104. doi:10.2174/0929867325666180111100601. PMID 29332565.
- 1 2 3 4 5 6 7 8 9 Pike, L. J. (2008). "The challenge of lipid rafts". The Journal of Lipid Research. 50 (Suppl): S323–8. doi:10.1194/jlr.R800040-JLR200. PMC 2674732. PMID 18955730.
- ↑ Simons, Kai; Ehehalt, Robert (2002). "Cholesterol, lipid rafts, and disease". Journal of Clinical Investigation. 110 (5): 597–603. doi:10.1172/JCI16390. PMC 151114. PMID 12208858.
- ↑ Laura, Anchisi; Sandra Dessi; Alessandra Pani; Antonella Mandas (25 November 2012). "Cholesterol homeostasis: a key to prevent or slow down neurodegeneration". Frontiers in Physiology. 3: 486. doi:10.3389/fphys.2012.00486. PMC 3539713. PMID 23316166.
- 1 2 Fantini, Jacques; Garmy, Nicolas; Mahfoud, Radhia; Yahi, Nouara (2004). "Lipid rafts: Structure, function and role in HIV, Alzheimer's and prion diseases". Expert Reviews in Molecular Medicine. 4 (27): 1–22. doi:10.1017/S1462399402005392. PMID 14987385. S2CID 2638429.
- ↑ Rietveld, Anton; Simons, Kai (1998). "The differential miscibility of lipids as the basis for the formation of functional membrane rafts". Biochimica et Biophysica Acta (BBA) - Reviews on Biomembranes. 1376 (3): 467–79. doi:10.1016/S0304-4157(98)00019-7. PMID 9805010.
- ↑ Radeva, Galina; Sharom, Frances J. (2004-05-15). "Isolation and characterization of lipid rafts with different properties from RBL-2H3 (rat basophilic leukaemia) cells". Biochemical Journal. 380 (1): 219–230. doi:10.1042/bj20031348. ISSN 0264-6021. PMC 1224147. PMID 14769131.
- ↑ Fivaz, Marc; Abrami, Laurence; Van Der Goot, F.Gisou (1999). "Landing on lipid rafts". Trends in Cell Biology. 9 (6): 212–3. doi:10.1016/S0962-8924(99)01567-6. PMID 10354632.
- ↑ Heerklotz, H. (2002). "Triton Promotes Domain Formation in Lipid Raft Mixtures". Biophysical Journal. 83 (5): 2693–701. Bibcode:2002BpJ....83.2693H. doi:10.1016/S0006-3495(02)75278-8. PMC 1302353. PMID 12414701.
- ↑ Levental, I; Lingwood, D; Grzybek, M; Coskun, U; Simons, K (21 December 2010). "Palmitoylation regulates raft affinity for the majority of integral raft proteins". Proceedings of the National Academy of Sciences of the United States of America. 107 (51): 22050–4. Bibcode:2010PNAS..10722050L. doi:10.1073/pnas.1016184107. PMC 3009825. PMID 21131568.
- ↑ Petersen, EN; Chung, HW; Nayebosadri, A; Hansen, SB (15 December 2016). "Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D." Nature Communications. 7: 13873. Bibcode:2016NatCo...713873P. doi:10.1038/ncomms13873. PMC 5171650. PMID 27976674.
- ↑ Robinson, CV; Rohacs, T; Hansen, SB (September 2019). "Tools for Understanding Nanoscale Lipid Regulation of Ion Channels". Trends in Biochemical Sciences. 44 (9): 795–806. doi:10.1016/j.tibs.2019.04.001. PMC 6729126. PMID 31060927.
- ↑ Singer, S. J.; Nicolson, Garth L. (1972). "The Fluid Mosaic Model of the Structure of Cell Membranes". Science. 175 (4023): 720–31. Bibcode:1972Sci...175..720S. doi:10.1126/science.175.4023.720. PMID 4333397. S2CID 83851531.
- ↑ Stier, A.; Sackmann, E. (1973). "Spin labels as enzyme substrates Heterogeneous lipid distribution in liver microsomal membranes". Biochimica et Biophysica Acta (BBA) - Biomembranes. 311 (3): 400–8. doi:10.1016/0005-2736(73)90320-9. PMID 4354130.
- ↑ Karnovsky, Morris J.; Kleinfeld, Alan M.; Hoover, Richard L.; Klausner, Richard D. (1982). "The concept of lipid domains in membranes". The Journal of Cell Biology. 94 (1): 1–6. doi:10.1083/jcb.94.1.1. PMC 2112185. PMID 6889603.
- ↑ Israelachvili, J. N.; Marčelja, S.; Horn, R. G. (2009). "Physical principles of membrane organization". Quarterly Reviews of Biophysics. 13 (2): 121–200. doi:10.1017/S0033583500001645. hdl:10536/DRO/DU:30041475. PMID 7015403. S2CID 33387523.
- ↑ Estep, T. N.; Mountcastle, D. B.; Barenholz, Y.; Biltonen, R. L.; Thompson, T. E. (1979). "Thermal behavior of synthetic sphingomyelin-cholesterol dispersions". Biochemistry. 18 (10): 2112–7. doi:10.1021/bi00577a042. PMID 435470.
- ↑ Goodsaid-Zalduondo, F.; Rintoul, D. A.; Carlson, J. C.; Hansel, W. (1982). "Luteolysis-Induced Changes in Phase Composition and Fluidity of Bovine Luteal Cell Membranes". Proceedings of the National Academy of Sciences of the United States of America. 79 (14): 4332–6. Bibcode:1982PNAS...79.4332G. doi:10.1073/pnas.79.14.4332. JSTOR 12587. PMC 346665. PMID 6956862.
- ↑ Simons, Kai; Van Meer, Gerrit (1988). "Lipid sorting in epithelial cells". Biochemistry. 27 (17): 6197–202. doi:10.1021/bi00417a001. hdl:1874/293951. PMID 3064805.
- ↑ Simons, Kai; Ikonen, Elina (1997). "Functional rafts in cell membranes". Nature. 387 (6633): 569–72. Bibcode:1997Natur.387..569S. doi:10.1038/42408. PMID 9177342. S2CID 4359503.
- 1 2 3 4 Allen, John A.; Halverson-Tamboli, Robyn A.; Rasenick, Mark M. (2006). "Lipid raft microdomains and neurotransmitter signalling". Nature Reviews Neuroscience. 8 (2): 128–40. doi:10.1038/nrn2059. PMID 17195035. S2CID 2098892.
- 1 2 Janes, Peter W.; Ley, Steven C.; Magee, Anthony I.; Kabouridis, Panagiotis S. (2000). "The role of lipid rafts in T cell antigen receptor (TCR) signalling". Seminars in Immunology. 12 (1): 23–34. doi:10.1006/smim.2000.0204. PMID 10723795.
- ↑ Schmitz, Gerd; Grandl, Margot (2008). "Update on lipid membrane microdomains". Current Opinion in Clinical Nutrition and Metabolic Care. 11 (2): 106–12. doi:10.1097/MCO.0b013e3282f44c2c. PMID 18301084. S2CID 13102246. Archived from the original on 2022-04-22. Retrieved 2019-11-24.
- ↑ Miljan, Erik A; Bremer, Eric G. (2002). "Regulation of growth factor receptors by gangliosides". Sci STKE. 2002 (160): RE15. doi:10.1126/stke.2002.160.re15. PMID 12454318. S2CID 84509111.
- ↑ Coskun, Ünal; Grzybek, Michal; Drechsel, David; Simons, Kai (2011). "Regulation of human EGF receptor by lipids". Proc Natl Acad Sci U S A. 108 (22): 9044–8. Bibcode:2011PNAS..108.9044C. doi:10.1073/pnas.1105666108. PMC 3107302. PMID 21571640.
- 1 2 Kovács, Tamás; Batta, Gyula; Zákány, Florina; Szöllősi, János; Nagy, Peter (2017). "The dipole potential correlates with lipid raft markers in the plasma membrane of living cells". J Lipid Res. 58 (8): 1681–1691. doi:10.1194/jlr.M077339. PMC 5538289. PMID 28607008.
- 1 2 3 Kovács, Tamás; Batta, Gyula; Hajdu, Tímea; Szabó, Ágnes; Váradi, Tímea; Zákány, Florina; Csomós, István; Szöllősi, János; Nagy, Peter (2016). "The Dipole Potential Modifies the Clustering and Ligand Binding Affinity of ErbB Proteins and Their Signaling Efficiency". Sci Rep. 6: 35850. Bibcode:2016NatSR...635850K. doi:10.1038/srep35850. PMC 5075772. PMID 27775011.
- ↑ Roepstorff, Kirstine; Thomsen, Peter; Sandvig, Kirsten; van Deurs, Deurs (2002). "Sequestration of epidermal growth factor receptors in non-caveolar lipid rafts inhibits ligand binding". J Biol Chem. 277 (21): 18954–60. doi:10.1074/jbc.M201422200. PMID 11886870.
- ↑ Mineo, Chieko; Gill, Gordon N.; Anderson, Richard G.W. (1999). "Regulated migration of epidermal growth factor receptor from caveolae". J Biol Chem. 274 (43): 30636–43. doi:10.1074/jbc.274.43.30636. PMID 10521449.
- ↑ Lambert, Sylviane; Vind-Kezunovic, Dina; Karvinen, Susanna; Gniadecki, Robert (May 2006). "Ligand-Independent Activation of the EGFR by Lipid Raft Disruption". Journal of Investigative Dermatology. 126 (5): 954–962. doi:10.1038/sj.jid.5700168. PMID 16456534.
- ↑ Nagy, Peter; Vereb, György; Sebestyén, Zsolt; Horváth, Gábor; Lockett, Stephen J.; Damjanovich, Sándor; Park, John W.; Jovin, Thomas M.; Szöllősi, János (2002-11-15). "Lipid rafts and the local density of ErbB proteins influence the biological role of homo- and heteroassociations of ErbB2". Journal of Cell Science. 115 (22): 4251–4262. doi:10.1242/jcs.00118. hdl:2437/80814. ISSN 0021-9533. PMID 12376557.
- ↑ Hofman, Erik G.; Ruonala, Mika O.; Bader, Arjen N.; Heuvel, Dave van den; Voortman, Jarno; Roovers, Rob C.; Verkleij, Arie J.; Gerritsen, Hans C.; Henegouwen, Paul M. P. van Bergen en (2008-08-01). "EGF induces coalescence of different lipid rafts". Journal of Cell Science. 121 (15): 2519–2528. doi:10.1242/jcs.028753. ISSN 0021-9533. PMID 18628305.
- ↑ FILIPP, D (2004). "Lipid rafts: resolution of the ?fyn problem??". Molecular Immunology. 41 (6–7): 645–656. doi:10.1016/j.molimm.2004.04.011. PMID 15220001.
- ↑ Irwin, Mary E.; Mueller, Kelly L.; Bohin, Natacha; Ge, Yubin; Boerner, Julie L. (2011-09-01). "Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib". Journal of Cellular Physiology. 226 (9): 2316–2328. doi:10.1002/jcp.22570. ISSN 1097-4652. PMC 3103760. PMID 21660955.
- 1 2 3 Field, Kenneth A.; Holowka, David; Baird, Barbara (1995). "FcɛRI-Mediated Recruitment of p53/56lyn to Detergent- Resistant Membrane Domains Accompanies Cellular Signaling". Proceedings of the National Academy of Sciences of the United States of America. 92 (20): 9201–5. Bibcode:1995PNAS...92.9201F. doi:10.1073/pnas.92.20.9201. JSTOR 2368438. PMC 40952. PMID 7568101.
- 1 2 Sheets, Erin D; Holowka, David; Baird, Barbara (1999). "Membrane organization in immunoglobulin E receptor signaling". Current Opinion in Chemical Biology. 3 (1): 95–9. doi:10.1016/S1367-5931(99)80017-9. PMID 10021405.
- 1 2 Baird, Barbara; Sheets, Erin D; Holowka, David (1999). "How does the plasma membrane participate in cellular signaling by receptors for immunoglobulin E?". Biophysical Chemistry. 82 (2–3): 109–19. doi:10.1016/S0301-4622(99)00110-6. PMID 10631794.
- ↑ Stauffer, Thomas P.; Meyer, Tobias (1997). "Compartmentalized IgE Receptor-mediated Signal Transduction in Living Cells". The Journal of Cell Biology. 139 (6): 1447–54. doi:10.1083/jcb.139.6.1447. PMC 2132626. PMID 9396750.
- ↑ Holowka, David; Sheets, Erin D.; Baird, Barbara (2000). "Interactions between FcεRI and lipid raft components are regulated by the actin cytoskeleton". Journal of Cell Science. 113 (6): 1009–19. doi:10.1242/jcs.113.6.1009. PMID 10683149.
- ↑ Sheets, E. D.; Holowka, D; Baird, B (1999). "Critical Role for Cholesterol in Lyn-mediated Tyrosine Phosphorylation of Fcepsilon RI and Their Association with Detergent-resistant Membranes". The Journal of Cell Biology. 145 (4): 877–87. doi:10.1083/jcb.145.4.877. PMC 2133197. PMID 10330413.
- ↑ Goitsuka, R.; Kanazashi, H; Sasanuma, H; Fujimura, Y; Hidaka, Y; Tatsuno, A; Ra, C; Hayashi, K; Kitamura, D (2000). "A BASH/SLP-76-related adaptor protein MIST/Clnk involved in IgE receptor-mediated mast cell degranulation". International Immunology. 12 (4): 573–80. doi:10.1093/intimm/12.4.573. PMID 10744659.
- ↑ Langlet, Claire; Bernard, Anne-Marie; Drevot, Philippe; He, Hai-Tao (2000). "Membrane rafts and signaling by the multichain immune recognition receptors". Current Opinion in Immunology. 12 (3): 250–5. doi:10.1016/S0952-7915(00)00084-4. PMID 10781401.
- ↑ Zhang, W; Trible, RP; Samelson, LE (1998). "LAT PalmitoylationIts Essential Role in Membrane Microdomain Targeting and Tyrosine Phosphorylation during T Cell Activation". Immunity. 9 (2): 239–46. doi:10.1016/S1074-7613(00)80606-8. PMID 9729044.
- ↑ Brdička, Tomáš; Černý, Jan; Hořejší, Václav (1998). "T Cell Receptor Signalling Results in Rapid Tyrosine Phosphorylation of the Linker Protein LAT Present in Detergent-Resistant Membrane Microdomains". Biochemical and Biophysical Research Communications. 248 (2): 356–60. doi:10.1006/bbrc.1998.8857. PMID 9675140. S2CID 782789.
- ↑ Cooper, Jonathan A.; Cary, Leslie A. (2000). "Molecular switches in lipid rafts". Nature. 404 (6781): 945–947. doi:10.1038/35010257. PMID 10801110.
- 1 2 Gupta, Neetu; Defranco, Anthony L. (2007). "Lipid rafts and B cell signaling". Seminars in Cell & Developmental Biology. 18 (5): 616–26. doi:10.1016/j.semcdb.2007.07.009. PMC 2169358. PMID 17719248.
- 1 2 3 4 Chazal, Nathalie; Gerlier, Denis (2003). "Virus Entry, Assembly, Budding, and Membrane Rafts". Microbiology and Molecular Biology Reviews. 67 (2): 226–37, table of contents. doi:10.1128/MMBR.67.2.226-237.2003. PMC 156468. PMID 12794191.
- 1 2 3 4 5 6 7 8 Pietiäinen, Vilja M.; Marjomäki, Varpu; Heino, Jyrki; Hyypiä, Timo (2005). "Viral entry, lipid rafts and caveosomes". Annals of Medicine. 37 (6): 394–403. doi:10.1080/07853890510011976. PMID 16203612. S2CID 40632761.
- ↑ Rajendran, Lawrence; Simons, Kai (2005). "Lipid rafts and membrane dynamics". Journal of Cell Science. 118 (6): 1099–102. doi:10.1242/jcs.01681. PMID 15764592.
- ↑ Rawat, Satinder S.; Viard, Mathias; Gallo, Stephen A.; Rein, Alan; Blumenthal, Robert; Puri, Anu (2003). "Modulation of entry of enveloped viruses by cholesterol and sphingolipids (Review)". Molecular Membrane Biology. 20 (3): 243–54. doi:10.1080/0968768031000104944. PMID 12893532. S2CID 36443978.
- ↑ Campbell, S.M; Crowe, S.M; Mak, J (2001). "Lipid rafts and HIV-1: From viral entry to assembly of progeny virions". Journal of Clinical Virology. 22 (3): 217–27. doi:10.1016/S1386-6532(01)00193-7. PMID 11564586.
- ↑ Alving, Carl R.; Beck, Zoltan; Karasavva, Nicos; Matyas, Gary R.; Rao, Mangala (2006). "HIV-1, lipid rafts, and antibodies to liposomes: Implications for anti-viral-neutralizing antibodies (Review)". Molecular Membrane Biology. 23 (6): 453–65. doi:10.1080/09687860600935348. PMID 17127618. S2CID 38887150.
- ↑ Wang, Hao; Yuan, Zixuan; Pavel, Mahmud Arif; Jablonski, Sonia Mediouni; Jablonski, Joseph; Hobson, Robert; Valente, Susana; Reddy, Chakravarthy B.; Hansen, Scott B. (10 May 2020). "The role of high cholesterol in age-related COVID19 lethality". bioRxiv 10.1101/2020.05.09.086249.
- ↑ Meng, B; et al. (March 2022). "Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity". Nature. 603 (7902): 706–714. Bibcode:2022Natur.603..706M. doi:10.1038/s41586-022-04474-x. PMC 8942856. PMID 35104837.
{{cite journal}}: CS1 maint: numeric names: authors list (link) - ↑ Yuan, Zixuan; Pavel, Mahmud Arif; Wang, Hao; Kwachukwu, Jerome C.; Mediouni, Sonia; Jablonski, Joseph Anthony; Nettles, Kendall W.; Reddy, Chakravarthy B.; Valente, Susana T.; Hansen, Scott B. (14 September 2022). "Hydroxychloroquine blocks SARS-CoV-2 entry into the endocytic pathway in mammalian cell culture". Communications Biology. 5 (1): 958. doi:10.1038/s42003-022-03841-8. PMC 9472185. PMID 36104427.
- ↑ Jacobson, Ken; Mouritsen, Ole G.; Anderson, Richard G. W. (2007). "Lipid rafts: At a crossroad between cell biology and physics". Nature Cell Biology. 9 (1): 7–14. doi:10.1038/ncb0107-7. PMID 17199125. S2CID 7876073.
- ↑ Sharma, Pranav; Varma, Rajat; Sarasij, R.C; Ira; Gousset, Karine; Krishnamoorthy, G; Rao, Madan; Mayor, Satyajit (2004). "Nanoscale Organization of Multiple GPI-Anchored Proteins in Living Cell Membranes". Cell. 116 (4): 577–89. doi:10.1016/S0092-8674(04)00167-9. PMID 14980224.
- ↑ Ritchie, Ken; Shan, Xiao-Yuan; Kondo, Junko; Iwasawa, Kokoro; Fujiwara, Takahiro; Kusumi, Akihiro (2005). "Detection of Non-Brownian Diffusion in the Cell Membrane in Single Molecule Tracking". Biophysical Journal. 88 (3): 2266–77. Bibcode:2005BpJ....88.2266R. doi:10.1529/biophysj.104.054106. PMC 1305276. PMID 15613635.
- ↑ Chiantia, Salvatore; et al. (2006). "Effects of ceramide on liquid-ordered domains investigated by simultaneous AFM and FCS". Biophysical Journal. 90 (12): 4500–4508. Bibcode:2006BpJ....90.4500C. doi:10.1529/biophysj.106.081026. PMC 1471841. PMID 16565041.
- ↑ Galvanetto, Nicola (2018). "Single-cell unroofing: probing topology and nanomechanics of native membranes". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1860 (12): 2532–2538. arXiv:1810.01643. Bibcode:2018arXiv181001643G. doi:10.1016/j.bbamem.2018.09.019. PMID 30273580. S2CID 52897823.
- ↑ Eggeling, Christian; Ringemann, Christian; Medda, Rebecca; Schwarzmann, Günter; Sandhoff, Konrad; Polyakova, Svetlana; Belov, Vladimir N.; Hein, Birka; et al. (2009). "Direct observation of the nanoscale dynamics of membrane lipids in a living cell". Nature. 457 (7233): 1159–62. Bibcode:2009Natur.457.1159E. doi:10.1038/nature07596. hdl:11858/00-001M-0000-0012-D8CA-4. PMID 19098897. S2CID 4428863.
- ↑ Thomas, S.; Kumar, R.S.; Casares, S.; Brumeanu, T.-D. (2003). "Sensitive detection of GM1 lipid rafts and TCR partitioning in the T cell membrane". Journal of Immunological Methods. 275 (1–2): 161–8. doi:10.1016/S0022-1759(03)00014-0. PMID 12667680.
- ↑ Thomas, Sunil; Kumar, Rajeev; Preda-Pais, Anca; Casares, Sofia; Brumeanu, Teodor-D. (2003). "A Model for Antigen-Specific T-Cell Anergy: Displacement of CD4-p56lck Signalosome from the Lipid Rafts by a Soluble, Dimeric Peptide-MHC Class II Chimera1". The Journal of Immunology. 170 (12): 5981–92. doi:10.4049/jimmunol.170.12.5981. PMID 12794125.
- ↑ Munro, Sean (2003). "Lipid rafts: elusive or illusive?". Cell. 115 (4): 377–88. doi:10.1016/S0092-8674(03)00882-1. PMID 14622593.
- ↑ Barenholz, Yechezkel (2004). "Sphingomyelin and Cholesterol: From Membrane Biophysics and Rafts to Potential Medical Applications". In Quinn, Peter J. (ed.). Membrane Dynamics and Domains. Subcellular Biochemistry. Vol. 37. pp. 167–215. doi:10.1007/978-1-4757-5806-1_5. ISBN 978-1-4419-3447-5. PMID 15376621.
- ↑ Pike, L. J.; Miller, JM (1998). "Cholesterol Depletion Delocalizes Phosphatidylinositol Bisphosphate and Inhibits Hormone-stimulated Phosphatidylinositol Turnover". Journal of Biological Chemistry. 273 (35): 22298–304. doi:10.1074/jbc.273.35.22298. PMID 9712847.
- ↑ Caroni, P. (2001). "NEW EMBO MEMBERS' REVIEW: Actin cytoskeleton regulation through modulation of PI(4,5)P2 rafts". The EMBO Journal. 20 (16): 4332–6. doi:10.1093/emboj/20.16.4332. PMC 125564. PMID 11500359.
- ↑ Kwik, Jeanne; Boyle, Sarah; Fooksman, David; Margolis, Leonid; Sheetz, Michael P.; Edidin, Michael (2003). "Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin". Proceedings of the National Academy of Sciences. 100 (24): 13964–9. Bibcode:2003PNAS..10013964K. doi:10.1073/pnas.2336102100. JSTOR 3148895. PMC 283529. PMID 14612561.
- ↑ Edidin, Michael (2003). "THE STATE OF LIPID RAFTS: From Model Membranes to Cells". Annual Review of Biophysics and Biomolecular Structure. 32: 257–83. doi:10.1146/annurev.biophys.32.110601.142439. PMID 12543707. S2CID 13122250.