Michael Travisano | |
|---|---|
 Michael Travisano in 2021 | |
| Born | February 12, 1961 |
| Alma mater | Columbia University Michigan State University |
| Scientific career | |
| Fields | Evolutionary biology |
| Institutions | University of Houston University of Minnesota Twin Cities |
Michael Travisano (born February 12, 1961) is an American evolutionary biologist, and a Distinguished McKnight University Professor at the University of Minnesota Twin Cities. In 2020, he started his position as Head Department in Ecology, Evolution & Behavior Department at the College of Biological Sciences.
Early life
Born in Nashville, Tennessee, Travisano is the son of Neal Travisano and Jo Anne Scriffiano. At the age of two he moved to Newark, New Jersey and remain there until 1969. He earned his Astrophysics BA from Columbia University in 1983.[1] Later in 1993, he obtained his PhD in Zoology from the Michigan State University.
Early career
Since 1983 he worked as laboratory technician in Charles Geard Radiology Research, Columbia University Physicians and Sciences. From 1986 to 1987 in Les Redpath Radiology, UC-Irvine. And from 1987 to 1988 at Richard Lenski laboratory. In 1993, he started his postdoctoral fellowship at the RIKEN Institute until 1994 in Saitama, Japan. Three years later from 1997 to 1999, Travisano did his second postdoctoral research at Oxford University, in the Department of Plant Sciences.
In 1999, he accepted a position as Assistant Professor at the University of Houston, where he was promoted to Associate Professor in 2006.
10,000 generations of E. coli
During his PhD at Michigan State University, Travisano worked in the long-term E. coli evolution experiment.[2] in Richard Lenski lab, following the evolutionary change in 12 populations of Escherichia coli propagated in 10,000 generations in identical environments. This works suggests chance events, such as mutation and drift, play an important role in adaptive evolution, as do the complex genetic interactions that underlie the structure of organisms.[2]
Academic work
His research mainly focus on experimental evolution, ecology and origins of life using microorganisms as models. The techniques of experimental evolution exploit the short-generation times of microbes to observe evolution in action, and to test explicit hypotheses about the effects of environmental manipulations on these processes. One main topic within experimental evolution research is the origin of multicellularity and its traits.
Although his primary appointment is in EEB he is also: 1) a member of the Biotechnology Institute; 2) the graduate program in microbial engineering; 3) the graduate program in plant and microbial biology; and 4) a resident fellow in the Minnesota Center for the Philosophy of Science.
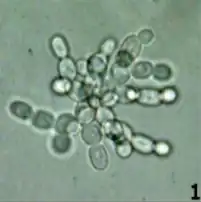
Multicellularity
The evolution of multicellularity is arguably the most significant innovation in the history of life after the origin of life itself.[3] The Travisano group showed that settling in a static test tube provided a simple selection scheme that favored the formation of multicellular clonal clusters in yeast--dubbed ‘snowflakes’ . Early multicellular clusters were composed of physiologically similar cells, but these subsequently evolved higher rates of programmed cell death, as is seen in the protective boundary of skin cells. In snowflake yeast, programmed cell death is an adaptation that increased cluster production.[3]
Niceness
Genes compete with one another for representation in the next generation, and the competitive nature of this process would seem to disfavor cooperation and niceness. Cells of brewer’s yeast release an enzyme that breaks indigestibly large sugar molecules into smaller, more easily digestible subunits. These digestible subunits are available to any yeast cell in the neighborhood, and the enzyme is costly, so surely selection should favor cheaters who chow down on the sugar subunits but don’t secrete the costly enzyme. Greig & Travisano, showed that selection for and against these cheaters depended on population size. Cheaters persist when populations are large, and when many ‘nice enzyme-secreting’ enzyme-secreting’ cells are around, but selection acts against cheaters when populations are low.[4]
.tif.jpg.webp)
Speciation
The mechanisms through which this separation is achieved are clearly fundamental to our understanding of the diversity of living things, since species are the raw material of organic diversity. Working with his postdoctoral associate, Duncan Greig, Travisano experimentally demonstrated speciation in the laboratory via a previously unknown mechanism. Publishing in Science, they reported that when a hybrid strain of yeast self-fertilizes its offspring are incompatible with either parent species but they produce fertile offspring when mated to each other: generating what is effectively an instant reproductively isolated species.[5]
Adaptive radiation
The Travisano-Rainey studies showed that in a matter of days, a single bacterium of Pseudomonas fluorescens will reproduce and evolve into three distinct lineages: one colonizes the air-water interface by forming a mat, one colonizes the bulk medium and one colonizes the anoxic environment at the bottom of the test tube. This happens if the test tube is unshaken, but not in a shaken test tube: demonstrating that environmental heterogeneity (like the different habitats of different islands) is key to the process of adaptive radiation[6]
References
- ↑ "Class of 1983". Columbia College Report. Retrieved 2022-08-13.
- 1 2 Lenski, R. E.; Travisano, M. (1994-07-19). "Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations". Proceedings of the National Academy of Sciences. 91 (15): 6808–6814. Bibcode:1994PNAS...91.6808L. doi:10.1073/pnas.91.15.6808. ISSN 0027-8424. PMC 44287. PMID 8041701.
- 1 2 Ratcliff, William C.; Denison, R. Ford; Borrello, Mark; Travisano, Michael (2012-01-31). "Experimental evolution of multicellularity". Proceedings of the National Academy of Sciences. 109 (5): 1595–1600. Bibcode:2012PNAS..109.1595R. doi:10.1073/pnas.1115323109. ISSN 0027-8424. PMC 3277146. PMID 22307617.
- ↑ Greig, Duncan; Travisano, Michael (2004-02-07). "The Prisoner's Dilemma and polymorphism in yeast SUC genes". Proceedings of the Royal Society of London. Series B: Biological Sciences. 271 (suppl_3): S25–S26. doi:10.1098/rsbl.2003.0083. PMC 1810003. PMID 15101409.
- ↑ Greig, Duncan; Louis, Edward J.; Borts, Rhona H.; Travisano, Michael (2002-11-29). "Hybrid speciation in experimental populations of yeast". Science. 298 (5599): 1773–1775. Bibcode:2002Sci...298.1773G. doi:10.1126/science.1076374. ISSN 1095-9203. PMID 12459586. S2CID 29972396.
- ↑ Rainey, Paul B.; Travisano, Michael (July 1998). "Adaptive radiation in a heterogeneous environment". Nature. 394 (6688): 69–72. Bibcode:1998Natur.394...69R. doi:10.1038/27900. ISSN 1476-4687. PMID 9665128. S2CID 40896184.