In mathematics, the Milstein method is a technique for the approximate numerical solution of a stochastic differential equation. It is named after Grigori N. Milstein who first published it in 1974.[1][2]
Description
Consider the autonomous Itō stochastic differential equation:
with initial condition , where stands for the Wiener process, and suppose that we wish to solve this SDE on some interval of time . Then the Milstein approximation to the true solution is the Markov chain defined as follows:
- partition the interval into equal subintervals of width :
- set
- recursively define for by: where denotes the derivative of with respect to and:are independent and identically distributed normal random variables with expected value zero and variance . Then will approximate for , and increasing will yield a better approximation.
Note that when , i.e. the diffusion term does not depend on , this method is equivalent to the Euler–Maruyama method.
The Milstein scheme has both weak and strong order of convergence, , which is superior to the Euler–Maruyama method, which in turn has the same weak order of convergence, , but inferior strong order of convergence, .[3]
Intuitive derivation
For this derivation, we will only look at geometric Brownian motion (GBM), the stochastic differential equation of which is given by:
with real constants and . Using Itō's lemma we get:
Thus, the solution to the GBM SDE is:
where
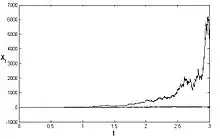
See numerical solution is presented above for three different trajectories.[4]
Computer implementation
The following Python code implements the Milstein method and uses it to solve the SDE describing the Geometric Brownian Motion defined by
# -*- coding: utf-8 -*-
# Milstein Method
import numpy as np
import matplotlib.pyplot as plt
class Model:
"""Stochastic model constants."""
μ = 3
σ = 1
def dW(Δt):
"""Random sample normal distribution."""
return np.random.normal(loc=0.0, scale=np.sqrt(Δt))
def run_simulation():
""" Return the result of one full simulation."""
# One second and thousand grid points
T_INIT = 0
T_END = 1
N = 1000 # Compute 1000 grid points
DT = float(T_END - T_INIT) / N
TS = np.arange(T_INIT, T_END + DT, DT)
Y_INIT = 1
# Vectors to fill
ys = np.zeros(N + 1)
ys[0] = Y_INIT
for i in range(1, TS.size):
t = (i - 1) * DT
y = ys[i - 1]
dw = dW(DT)
# Sum up terms as in the Milstein method
ys[i] = y + \
Model.μ * y * DT + \
Model.σ * y * dw + \
(Model.σ**2 / 2) * y * (dw**2 - DT)
return TS, ys
def plot_simulations(num_sims: int):
"""Plot several simulations in one image."""
for _ in range(num_sims):
plt.plot(*run_simulation())
plt.xlabel("time (s)")
plt.ylabel("y")
plt.grid()
plt.show()
if __name__ == "__main__":
NUM_SIMS = 2
plot_simulations(NUM_SIMS)
See also
References
- ↑ Mil'shtein, G. N. (1974). "Approximate integration of stochastic differential equations". Teoriya Veroyatnostei i ee Primeneniya (in Russian). 19 (3): 583–588.
- ↑ Mil’shtein, G. N. (1975). "Approximate Integration of Stochastic Differential Equations". Theory of Probability & Its Applications. 19 (3): 557–000. doi:10.1137/1119062.
- ↑ V. Mackevičius, Introduction to Stochastic Analysis, Wiley 2011
- ↑ Umberto Picchini, SDE Toolbox: simulation and estimation of stochastic differential equations with Matlab. http://sdetoolbox.sourceforge.net/
Further reading
- Kloeden, P.E., & Platen, E. (1999). Numerical Solution of Stochastic Differential Equations. Springer, Berlin. ISBN 3-540-54062-8.
{{cite book}}: CS1 maint: multiple names: authors list (link)