| NBPF26 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | NBPF26, NBPF member 26 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | HomoloGene: 41035 GeneCards: NBPF26 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
NBPF26, or Neuroblastoma breakpoint family member 26, is a protein encoded by the NBPF26 gene in Homo sapiens. The alias for NBPF26 is notch 2 N-terminal like R (NOTCH2NLR).[3] NBPF26 encodes 13 Olduvai domains, which are thought to contribute to the rapid expansion of the neocortex in humans.[4]
Gene
Locus
The NBPF26 gene is located on the plus strand of chromosome 1 (1p11.2) from 120,723,945 to 120,842,229, spanning 118,285 base pairs. NOTCH2NLR is an alias of NBPF26 and overlaps with beginning of NBPF26's coding sequence.
Genomic neighborhood

Within the genomic neighborhood of human NBPF26, phosphodiesterase 4D interacting protein-like pseudogene 2 (PDE4DIPP2), NOTCH2NLR, Rosellinia necatrix victorivirus 1-19 (RnVV1-19), and tRNA-Asn (anticodon GTT) 7-1 (TRN-GTT7-1) can be found.
mRNA
NBPF26 encodes three isoform variants shown in the table below. Isoform one is the longest variant and spans 6,891 base pairs.[5]
| Isoform | Nucleotide Accession # | mRNA Length (bp) | Protein Accession # | Protein Length (aa) | Molecular Weight (kDa) |
|---|---|---|---|---|---|
| 1 | NM_001405520.1 | 6,891 | NP_001392449.1 | 1,673 | 190 |
| 2 | NM_001351372.2 | 6,666 | NP_001338301.2 | 1,598 | 182 |
| 3 | NM_001395637.2 | 6,216 | NP_001382566.1 | 1448 | 165 |
Protein
Composition
NBPF26 isoform one has a predicted molecular weight of 190 kDa and an isoelectric point of 4.6.[7] Relative to other proteins, NBPF26 is rich in glutamic acid, glutamine, and cysteine, and poor in isoleucine.[8] Two positive charge clusters, spanning 22 amino acids, are located at positions 1,241-1,262 and 1,560-1,581.
Motifs
Five EGF-like domains and one calcium-binding EGF-like domain are encoded in NBPF26.[9]
NBPF26 also encodes 13 Olduvai domains.[10] Positions and sequences of Conserved 1-3 (Con1-3) and Human lineage-specific 1-3 (HLS1-3) Olduvai clades are shown in the table below.
| Clade | Position (aa) | Sequence |
|---|---|---|
| Con1 | 442-505 | EKVLESSAPREVQKTEESKVPEDSLEECAITCSNSHGPCDSNQPHKNIKITFEEDEVNSTLVVD |
| Con1 | 713-775 | EKVQKSSAPREMQKAEEKEVPEDSLEECAITCSNSHGPYDCNQPHRKTKITFEEDKVDSTLIG |
| Con2 | 799-862 | EEKGPVSPRNLQESEEEEVPQESWDEGYSTLSIPPEMLASYKSYSSTFHSLEEQQVCMAVDIG |
| HLS1 | 871-937 | KEDHEATGPRLSRELLDEKGPEVLQDSLDRCYSTPSGCLELTDSCQPYRSAFYVLEQQRVGLAVDMD |
| HLS1 | 946-1,012 | EEDQDPSCPRLSGELLDEKEPEVLQESLDRCYSTPSGCLELTDSCQPYRSAFYILEQQRVGLAVDMD |
| HLS1 | 1,021-1,087 | EEDQDPSCPRLSGELLDEKEPEVLQESLDRCYSTPSGCLELTDSCQPYRSAFYILEQQRVGLAVDMD |
| HLS2 | 1,096-1,162 | EEDQDPSCPRLSRELLDEKEPEVLQDSLGRCYSTPSGYLELPDLGQPYSSAVYSLEEQYLGLALDVD |
| HLS3 | 1,171-1,237 | EEDQGPPCPRLSRELLEVVEPEVLQDSLDRCYSTPSSCLEQPDSCQPYGSSFYALEEKHVGFSLDV |
| HLS1 | 1,265-1331 | EEDQNPPCPRLSRELLDEKGPEVLQDSLDRCYSTPSGCLELTDSCQPYRSAFYILEQQRVGLAVDMD |
| HLS3 | 1,340-1,406 | EEDQDPSCPRLSRELLEVVEPEVLQDSLDRCYSTPSSCLEQPDSCQPYGSSFYALEEKHVGFSLDVG |
| HLS2 | 1,415-1,481 | EEDQDPSCPRLSRELLDEKEPEVLQDSLGRCYSTPSGYLELPDLGQPYSSAVYSLEEQYLGLALDVD |
| HLS3 | 1,490-1,556 | EEDQGPPCPRLSRELLEVVEPEVLQDSLDRCYSTPSSCLEQPDSCQPYGSSFYALEEKHVGFSLDVG |
| Con3 | 1,584-1,650 | EEDQNPPCPRLNSMLMEVEEPEVLQDSLDICYSTPSMYFELPDSFQHYRSVFYSFEEEHISFALYVD |
Post-translational modifications
NBPF26 is phosphorylated at amino acids 46, 48, 97, and 228.[11][12] In addition, NBPF26 is predicted to undergo sulfonation, C-mannosylation, N-glycosylation, and O-glycosylation.[13][14][15][16] Three furin cleavage sites are found at amino acid positions 379, 1252, and 1571.[17]
Protein interactions
NBPF26 is predicted to interact with Estrogen receptor 1 (ESR1), APEX nuclease (multifunctional DNA repair enzyme) 1 (APEX1), lysine (K)-specific demethylase 1A (KDM1A), trans-golgi network protein 2 (TGOLN2), and tripartite motif containing 25 (TRIM25).[18][19]
Homology
Orthologs
Distant orthologs for human NBPF26 can be found in primates, birds, reptiles, amphibians, fish, and some invertebrates. Select orthologs are shown in the table below.
| Class | Genus and Species | Common name | Gene name | Accession # | Length (aa) | Identity (%) | Similarity (%) |
|---|---|---|---|---|---|---|---|
| Mammalia | Pan troglodytes | Chimpanzee | NBPF14 | XP_054958524 | 1,615 | 53.9 | 57.8 |
| Mammalia | Gorilla gorilla gorilla | Gorilla | NBPF12 | XP_055212742.1 | 2,428 | 54.3 | 56.0 |
| Mammalia | Mus musculus | House Mouse | NOTCH2 | NP_035058.2 | 2,473 | 16.0 | 21.2 |
| Mammalia | Enhydra lutris kenyoni | Northern Sea Otter | NBPF26 | XP_022351917.1 | 2,358 | 12.3 | 21.6 |
| Aves | Dryobates pubescens | Downy woodpecker | NOTCH2 | XP_054021673.1 | 2,453 | 20.1 | 29.7 |
| Aves | Antrostomus carolinensis | Chuck-will's-widow | NOTCH2 | XP_010174161 | 2,401 | 19.1 | 28.5 |
| Aves | Strigops habroptila | Owl Parrot | NOTCH2 | XP_030350503.1 | 2,432 | 15.6 | 22.4 |
| Aves | Anser cygnoides | Swan goose | NBPF26 | XP_047936201.1 | 2,359 | 13.7 | 24.1 |
| Reptilia | Trachemys scripta elegans | Red-eared slider | NOTCH2 | XP_034634874.1 | 2,397 | 20.0 | 29.8 |
| Reptilia | Alligator mississippiensis | American alligator | NOTCH2 | XP_006260017.1 | 2,435 | 16.7 | 24.2 |
| Reptilia | Podarcis raffonei | Aeolian wall lizard | NOTCH1 | XP_053230981.1 | 2,531 | 14.6 | 22.8 |
| Reptilia | Pseudonaja textilis | Eastern brown snake | NOTCH3 | XP_026552021.1 | 2,416 | 14.3 | 21.3 |
| Amphibia | Spea bombifrons | Plains spadefoot toad | NOTCH2L | XP_053317909.1 | 2,419 | 17.0 | 25.7 |
| Amphibia | Xenopus laevis | African clawed frog | NOTCH2 | XP_018114267.1 | 2,450 | 15.0 | 22.6 |
| Amphibia | Geotrypetes seraphini | Gaboon caecilian | NOTCH3 | XP_033779639.1 | 2,472 | 14.6 | 21.8 |
| Amphibia | Hyla sarda | Sardinian tree frog | MTF1 | XP_056404323.1 | 1,169 | 12.4 | 22.3 |
| Chordata | Acipenser ruthenus | Sterlet | NOTCH2 | XP_058887354.1 | 2,454 | 16.4 | 24.7 |
| Chordata | Callorhinchus milii | Australian ghostshark | NOTCH2 | XP_042190840.1 | 2,819 | 16.0 | 23.8 |
| Chordata | Petromyzon marinus | Sea lamprey | NOTCH1L | XP_032815028.1 | 2,536 | 14.9 | 23.2 |
| Chordata | Paramormyrops kingsleyae | Elephantfish | NOTCH2L | XP_023699719.1 | 2,420 | 14.4 | 22.2 |
| Invertebrates | Amphimedon queenslandica | Sponge | FBPP1L | XP_019855855 | 698 | 8.1 | 14.4 |
Paralogs
Human NBPF26 has multiple paralogs. The top 10 are shown in the table below.
| Name | Accession # | Length (aa) | Identity (%) | Similarity (%) |
|---|---|---|---|---|
| NBPF1 | NP_001392595.1 | 1,704 | 68.8 | 71.6 |
| NBPF7 | NP_001392671.1 | 1,704 | 68.2 | 71.0 |
| LOC102724250 | NP_001392459.1 | 1,704 | 67.7 | 70.4 |
| NBPF1L | NP_001393481.1 | 1,949 | 66.1 | 68.7 |
| NBPF12 | KAI4082365.1 | 1,459 | 65.1 | 67.3 |
| NBPF9 | NP_001264373.1 | 1,111 | 64.7 | 65.2 |
| NBPF20 | XP_047301971.1 | 1,065 | 61.7 | 62.5 |
| KIAA1693 | BAB21784.1 | 901 | 50.7 | 51.8 |
| NBPF8 | XP_047285792.1 | 1,353 | 48.6 | 50.0 |
| NBPF11 | NP_001095133.3 | 865 | 46.3 | 47.9 |
Expression
NBPF26 RNA expression is ubiquitous throughout human tissue.[20]
Clinical Significance
In lung cancer, mutations in NBPF26 were associated with local disease recurrence.[21] NBPF26 expression in monocytes was increased in pregnant patients with rheumatoid arthritis compared to pregnancies without rheumatoid arthritis.[22]
Conceptual translation
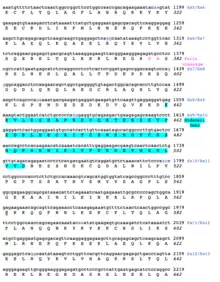
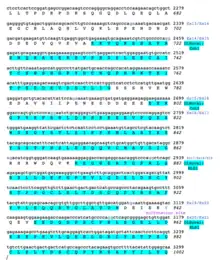
A conceptual translation of human NBPF26 is shown below.

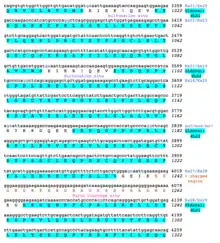
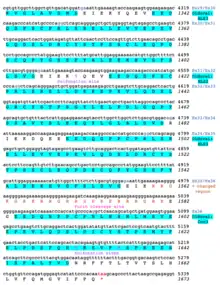

References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000273136 - Ensembl, May 2017
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ NCBI gene entry for NBPF26
- ↑ Fiddes, I. T., Pollen, A. A., Davis, J. M., & Sikela, J. M. (2019). Paired involvement of human-specific Olduvai domains and NOTCH2NL genes in human brain evolution. Human genetics, 138, 715-721.
- ↑ NCBI gene entry for NBPF26
- ↑ AlphaFold <https://alphafold.ebi.ac.uk/entry/A0A2J8IZQ5>
- ↑ Expasy compute pI <https://web.expasy.org/cgi-bin/compute_pi/pi_tool>
- ↑ Statistical Analysis of Protein Sequences (SAPS)<https://www.ebi.ac.uk/Tools/seqstats/saps>
- ↑ MotifFinder <https://www.genome.jp/tools-bin/search_motif_lib>
- ↑ MyHits Motif Scan <https://myhits.sib.swiss/cgi-bin/motif_scan>
- ↑ DTU NetPhos <https://services.healthtech.dtu.dk/services/NetPhos-3.1/>
- ↑ GPS <https://gps.biocuckoo.cn/online.php>
- ↑ Sulfinator <https://web.expasy.org/sulfinator/>
- ↑ NetCGlyc <https://services.healthtech.dtu.dk/services/NetCGlyc-1.0/>
- ↑ NetNGlyc 1.0 < https://services.healthtech.dtu.dk/services/NetNGlyc-1.0/>
- ↑ NetOGlyc 4.0 <https://services.healthtech.dtu.dk/services/NetOGlyc-4.0/>
- ↑ ProP 1.0 <https://services.healthtech.dtu.dk/services/ProP-1.0/>
- ↑ IntAct database <https://www.ebi.ac.uk/intact/home>
- ↑ BioGrid < https://thebiogrid.org/>
- ↑ NBPF26 RNA expression <https://www.ncbi.nlm.nih.gov/gene/101060684>
- ↑ Valter, A., Luhari, L., Pisarev, H., Truumees, B., Planken, A., Smolander, O. P., & Oselin, K. (2023). Genomic alterations as independent prognostic factors to predict the type of lung cancer recurrence. Gene, 885, 147690. https://doi.org/10.1016/j.gene.2023.147690
- ↑ Lien, H. J. T., Pedersen, T. T., Jakobsen, B., Flatberg, A., Chawla, K., Sætrom, P., & Fenstad, M. H. (2023). Single-cell resolution of longitudinal blood transcriptome profiles in rheumatoid arthritis, systemic lupus erythematosus and healthy control pregnancies. Annals of the rheumatic diseases, ard-2023-224644. Advance online publication. https://doi.org/10.1136/ard-2023-224644