| Nickel superoxide dismutase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
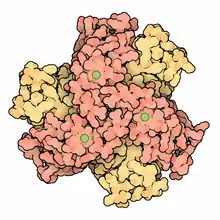 Structure of Streptomyces Ni superoxide dismutase hexamer[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Sod_Ni | ||||||||
| Pfam | PF09055 | ||||||||
| InterPro | IPR014123 | ||||||||
| SCOP2 | 1q0d / SCOPe / SUPFAM | ||||||||
| |||||||||
Nickel superoxide dismutase (Ni-SOD) is a metalloenzyme that, like the other superoxide dismutases, protects cells from oxidative damage by catalyzing the disproportionation of the cytotoxic superoxide radical (O−
2) to hydrogen peroxide and molecular oxygen. Superoxide is a reactive oxygen species that is produced in large amounts during photosynthesis and aerobic cellular respiration.[2] The equation for the disproportionation of superoxide is shown below:
Ni-SOD was first isolated in 1996 from Streptomyces bacteria and is primarily found in prokaryotic organisms. It has since been observed in cyanobacteria and a number of other aquatic microbes.[3] Ni-SOD is homohexameric, meaning that it has six identical subunits. Each subunit has a single nickel containing active site.[4] The disproportionation mechanism involves a reduction-oxidation cycle where a single electron transfer is catalyzed by the Ni2+/Ni3+ redox couple.[4][2] Ni-SOD catalyzes close to the barrier of diffusion.
Structure
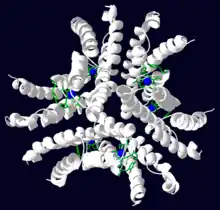
Ni-SOD is a globular protein and is shaped like a hollow sphere. It is homohexameric, meaning that it is made up of six identical subunits. Each subunit is a bundle of four right-handed α-helixes and has a molecular mass of 13.4 kDa (117 amino acids).[5] The subunits align to give Ni-SOD a three-fold axis of symmetry.[1] There are six nickel cofactors in total (one for each subunit). The subunits also have a hydrophobic core, which helps drive protein folding. The core is made up of 17 aliphatic amino acids.
Nickel binding hook
All of the amino acids involved in catalysis and nickel binding are located within the first six residues from the N-terminus of each subunit.[4][2] This region has a curved and disordered shape in the absence of nickel, which gives it its nickname, “the nickel binding hook”. After nickel binds, this motif takes on a highly ordered structure and forms the enzyme's active site. The nickel binding hook is composed of the conserved sequence H2N-His-Cys-X-X-Pro-Cys-Gly-X-Tyr (where X could be any amino acid, i.e. the position isn't conserved).[5] Proline-5 creates a sharp turn, giving this region a hook shape.[1][5] His-1, Cys-2, Cys-6 and the N-terminus make up the ligand set for the nickel cofactors. After nickel binds the ordered structure of the nickel binding hook is stabilized by forming hydrogen bonds with amino acids at the interface of two different subunits.[1]
Active site
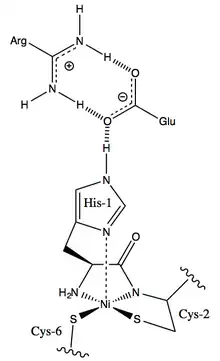
The six active sites are located in the nickel binding hook of each subunit.[4]
Ni-SOD is the only superoxide dismutase with ligands other than histidine, aspartate or water. The amino acid residues that define the coordination sphere of Ni are cysteine-2, cysteine-6 and histidine-1. The equatorial ligands include the thiolates of cysteine-2 and cysteine-6, as well as a deprotonated backbone amide nitrogen and the N-terminal amine. This is one of the few examples of a backbone amide group acting as a metal ligand in a protein.[4][2]
The thiolate sulfur centers are susceptible to oxidative damage.[4][6]
Coordination geometry the nickel cofactor
In its oxidized (Ni(III)) state, the coordination geometry of nickel is square pyramidal.[7] The binding of histidine-1 as an axial ligand in the reduced enzyme is uncertain.[3][7] If histidine is not a ligand in the reduced enzyme, the nickel(II) cofactor would be square planar.[4][5][7] However, the His-1 may remain in place throughout the redox cycle, meaning that the nickel cofactor would always have a square-pyramidal geometry. His-1 is held in place over the nickel cofactor in a tight hydrogen bonding network with a glutamic acid residue and an arginine residue.[4]
Mechanism
The catalytic mechanism is analogous to highly efficient "ping-pong" mechanism of copper-zinc superoxide dismutase, where O−
2 alternately reduces and oxidizes the nickel cofactor.[4][8] Two single electron transfer steps are involved:
Several aspects of the mechanism that are remain unclear. For example, both the H+ source and transfer mechanism are still vague. H+ is most likely carried into the active site by the substrate, meaning that superoxide enters the enzyme in its protonated form (HO2).[1][4][2] The disproportionation is most likely catalyzed in the second coordination sphere, but the mechanism of electron transfer is still up for debate.[4][2] It is possible that a quantum tunnelling event is involved.[4] Nickel superoxide dismutase is an incredibly efficient enzyme, indicating the redox mechanism is very fast. This means that large structural rearrangements or dramatic changes to the coordination sphere are unlikely to be involved in the catalytic mechanism.[4]
Occurrence
Nickel superoxide dismutase is primarily found in bacteria. The only known example of a eukaryote expressing a nickel containing superoxide dismutase is in the cytoplasm of a number of green algae species.[8] Ni-SOD was first isolated from Streptomyces bacteria, which are mostly found in soil. Streptomyces Ni-SOD has been the most heavily studied nickel containing SOD to date. These enzymes are now known to exist in a number of other prokaryotes, including cyanobacteria and several Actinomycetes species. Some of the Actinomycetes species that express nickel containing superoxide dismutatses are Micromonospora rosia, Microtetraspora glauca and Kitasatospora griseola.[1] Ni-SOD hasn't been found in any archaea.[1]
Regulation
Nickel is the primary regulatory factor in the expression of Ni-SOD. Increased nickel concentration in the cytosol increases the expression of sodN, the gene that encodes Ni-SOD in Streptomyces. In the absence of nickel sodN isn't transcribed, indicating that nickel positively regulates Ni-SOD expression. The folding of the enzyme is also dependent on the presence of nickel in the cytosol. As mentioned above, the nickel binding hook is disordered when nickel isn't present.
Nickel also acts as a negative regulator, repressing the transcription of other superoxide dismutases. In particular, expression of iron superoxide dismutase (Fe-SOD) is repressed by nickel in Streptomyces coelicolor.[9] A quintessential example of this negative regulation is Nur, nickel binding repressor.[3] When nickel is present Nur binds to the promoter of sodF, stopping the production of iron superoxide dismutase.
Post-translational modification is also required to produce the active enzyme. In order to expose the nickel-binding hook, a leader sequence must be enzymatically cleaved off the N-terminus.[3]
References
- 1 2 3 4 5 6 7 Wuerges J, Lee JW, Yim YI, Yim HS, Kang SO, Djinovic Carugo K (June 2004). "Crystal structure of nickel-containing superoxide dismutase reveals another type of active site". Proc. Natl. Acad. Sci. U.S.A. 101 (23): 8569–74. Bibcode:2004PNAS..101.8569W. doi:10.1073/pnas.0308514101. PMC 423235. PMID 15173586.
- 1 2 3 4 5 6 Pelmenschikov, Vladimir (2006). "Nickel Superoxide Dismutase Reaction Mechanism Studied by Hybrid Density Functional Methods". J. Am. Chem. Soc. 128 (23): 7466–7475. doi:10.1021/ja053665f. PMID 16756300.
- 1 2 3 4 Zamble, Deborah B.; Li, Yanjie (2009). "Nickel Homeostasis and Nickel Regulation: an Overview". Chemical Reviews. 109 (10): 4617–4643. doi:10.1021/cr900010n. PMID 19711977.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 Shearer, Jason (2014). "Insight into the Structure and Mechanism of Nickel-Containing Superoxide Dismutase Derived from Peptide-Based Mimics". Accounts of Chemical Research. 47 (8): 2332–2341. doi:10.1021/ar500060s. PMID 24825124.
- 1 2 3 4 5 Barondeau, David P. (2004). "Nickel Superoxide Dismutase Structure and Mechanism". Biochemistry. 43 (25): 8038–8047. doi:10.1021/bi0496081. PMID 15209499.
- ↑ Abreu, Isabel, A.; Cabelli, Diane, E. (2009). "Superoxide dismutateses-a review of the metal associated mechanistic variations". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1804 (2): 272. doi:10.1016/j.bbapap.2009.11.005. PMID 19914406.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - 1 2 3 Neupane, Kosh P.; Gearty, Kristie; Ashish, Francis; Shearer, Jason (2007). "Probing Variable Axial Ligation in Nickel Superoxide Dismutase Utilizing Metallopeptide-Based Models: Insight into the Superoxide Disproportionation Mechanism". J. Am. Chem. Soc. 129 (47): 14605–14618. doi:10.1021/ja0731625. PMID 17985883.
- 1 2 Sheng, Yuewei (2014). "Superoxide Dismutases and Superoxide Reductases". Chemical Reviews. 114 (7): 3854–3918. doi:10.1021/cr4005296. PMC 4317059. PMID 24684599.
- ↑ Mulrooney, Scott B.; Hausinger, Robert P. (2003). "Nickel uptake and utilization by microorganisms". FEMS Microbiology Reviews. 27 (2–3): 239–261. doi:10.1016/S0168-6445(03)00042-1. PMID 12829270.