A nuclear run-on assay is conducted to identify the genes that are being transcribed at a certain time point. Approximately one million cell nuclei are isolated and incubated with labeled nucleotides, and genes in the process of being transcribed are detected by hybridization of extracted RNA to gene specific probes on a blot.[1] Garcia-Martinez et al. (2004) [2] developed a protocol for the yeast S. cerevisiae (Genomic run-on, GRO) that allows for the calculation of transcription rates (TRs) for all yeast genes to estimate mRNA stabilities for all yeast mRNAs.[3]
Alternative microarray methods have recently been developed, mainly PolII RIP-chip: RNA immunoprecipitation of RNA polymerase II with phosphorylated C-terminal domain directed antibodies and hybridization on a microarray slide or chip (the word chip in the name stems from "ChIP-chip" where a special Affymetrix GeneChip was required). A comparison of methods based on run-on and ChIP-chip has been made in yeast (Pelechano et al., 2009). A general correspondence of both methods has been detected but GRO is more sensitive and quantitative. It has to be considered that run-on only detects elongating RNA polymerases whereas ChIP-chip detects all present RNA polymerases, including backtracked ones.
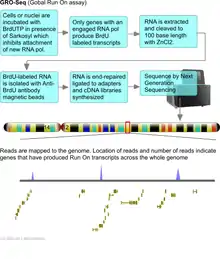
Attachment of new RNA polymerase to genes is prevented by inclusion of sarkosyl. Therefore only genes that already have an RNA polymerase will produce labeled transcripts. RNA transcripts that were synthesized before the addition of the label will not be detected as they will lack the label. These run on transcripts can also be detected by purifying labeled transcripts by using antibodies that detect the label and hybridizing these isolated transcripts with gene expression arrays or by next generation sequencing (GRO-Seq).[4]
Run on assays have been largely supplanted with Global Run on assays that use next generation DNA sequencing as a readout platform. These assays are known as GRO-Seq and provide an incredibly detailed view of genes engaged in transcription with quantitative levels of expression. Array based methods for analyzing Global run on (GRO) assays are being replaced with Next Generation Sequencing which eliminates the design of probes against gene sequences. Sequencing will catalog all transcripts produced even if they are not reported in databases. GRO-seq involves the labeling of newly synthesized transcripts with bromouridine (BrU). Cells or nuclei are incubated with BrUTP in the presence of sarkosyl, which prevents the attachment of RNA polymerase to the DNA. Therefore only RNA polymerase that are already on the DNA before the addition of sarkosyl will produce new transcripts that will be labeled with BrU. The labeled transcripts are captured with anti-BrU antibody labeled beads, converted to cDNAs and then sequenced by Next Generation DNA sequencing. The sequencing reads are then aligned to the genome and number of reads per transcript provide an accurate estimate of the number of transcripts synthesized.[5]
References
- ↑ Gariglio, P; Bellard, M; Chambon, P (Jun 1981). "Clustering of RNA polymerase B molecules in the 5' moiety of the adult beta-globin gene of hen erythrocytes". Nucleic Acids Res. 9 (11): 2589–98. doi:10.1093/nar/9.11.2589. PMC 326874. PMID 6269056.
- ↑ García-Martínez, J; Aranda, A; Pérez-Ortín, JE (2004). "Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms". Mol. Cell. 15 (2): 303–13. doi:10.1016/j.molcel.2004.06.004. hdl:10550/32058. PMID 15260981.
- ↑ Pelechano, V; Jimeno-González, S; Rodríguez-Gil, A; García-Martínez, J; Pérez-Ortín, JE; Chávez, S (Aug 2009). "Regulon-specific control of transcription elongation across the yeast genome". PLOS Genet. 5 (8): e1000614. doi:10.1371/journal.pgen.1000614. PMC 2721418. PMID 19696888.
- ↑ Core, L; Waterfall, J; Lis, JT (2008). "Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters". Science. 322 (5909): 1845–8. Bibcode:2008Sci...322.1845C. doi:10.1126/science.1162228. PMC 2833333. PMID 19056941.
- ↑ Min, IM; Waterfall, JJ; Core, LJ; Munroe, RJ; Schimenti, J; Lis, JT (Apr 2011). "Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells". Genes Dev. 25 (7): 742–54. doi:10.1101/gad.2005511. PMC 3070936. PMID 21460038.