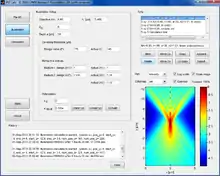
 PSF Lab on Windows 7 | |
| Developer(s) | Michael J. Nasse, Jörg C. Woehl |
|---|---|
| Stable release | 3.0
/ September 24, 2010 |
| Operating system | Windows, macOS |
| Type | Computer simulation software |
| License | Freeware for academic and non-commercial use |
| Website | onemolecule |
PSF Lab is a software program that allows the calculation of the illumination point spread function (PSF) of a confocal microscope under various imaging conditions. The calculation of the electric field vectors is based on a rigorous, vectorial model that takes polarization effects in the near-focus region and high numerical aperture microscope objectives into account.[1]
The polarization of the input beam (assumed to be collimated and monochromatic) can be chosen freely (linear, circular, or elliptic). Furthermore, a constant or Gaussian shaped input beam intensity profile can be assumed. On its way from the objective to the focus, the illumination light passes through up to three stratified optical layers, which allows the simulation of an immersion oil/air (layer 1) objective that focusses light through a glass cover slip (layer 2) into the sample medium (layer 3). Each layer is characterized by its (constant) refractive index and thickness. PSF Lab can also simulate microscope objectives that are corrected for certain refractive indices and cover slip thicknesses (design parameters). Thus, any deviations from the ideal imaging conditions for which the objective was designed for are properly taken into account.
The following optical parameters can be selected:[2]
- Input beam
- Wavelength
- Gaussian profile filling parameter (0 = constant profile)
- Polarization (linear, circular, elliptic)
- Outputs
- Individual field components
- Squared field components
- Intensity
- Microscope objective
- Numerical aperture
- Optical media
- Refractive index (design and actual)
- Thickness (design and actual)
- Depth (focus position within medium 3)
The program calculates only 2D section of the PSF, but several calculations can be stacked (with a third party program) to obtain the full 3D PSF. Calculations are organized in "sets", each with its own set of parameters. Loops can be set up such that PSF Lab calculates one or several sets, increasing the resolution of the calculated images in each new iteration. The resulting image is displayed in PSF Lab in linear or logarithmic color scale with user-selectable color map, and the intensity, individual field components, or squared field component distributions can be exported into various formats (data formats: .mat, .h5 (HDF5), .txt (ASCII); image formats: .fig, .ai, .bmp, .emf, .eps, .jpg, .pcx, .pdf, .png, .tif).
See also
References
- ↑ Nasse M. J., Woehl J. C. (2010). "Realistic modeling of the illumination point spread function in confocal scanning optical microscopy". J. Opt. Soc. Am. A. 27 (2): 295–302. Bibcode:2010JOSAA..27..295N. doi:10.1364/JOSAA.27.000295. PMID 20126241.
- ↑ PSF Lab Getting Started Manual
External links
- Official website
- Molecular Expressions, introduction to deconvolution using PSFs.