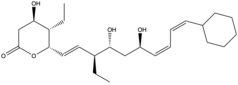
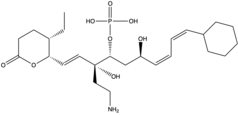
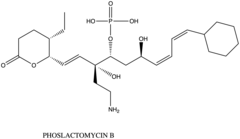
 | |
| Names | |
|---|---|
| Preferred IUPAC name
(1E,3R,4R,6R,7Z,9Z)-3-(2-Aminoethyl)-10-cyclohexyl-1-[(2S,3S)-3-ethyl-6-oxo-3,6-dihydro-2H-pyran-2-yl]-3,6-dihydroxydeca-1,7,9-trien-4-yl dihydrogen phosphate | |
| Other names
2H-Pyran-2-one, 6-(3-(2-aminoethyl)-10-cyclohexyl-3,6-dihydroxy-4-(phosphonooxy)-1,7,9-decatrienyl)-5-ethyl-5,6-dihydro- | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChemSpider | |
| KEGG | |
PubChem CID |
|
| |
| |
| Properties | |
| C25H40NO8P | |
| Molar mass | 513.568 g·mol−1 |
| Density | 1.3 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
Phoslactomycin (PLM) is a natural product from the isolation of Streptomyces species. This is an inhibitor of the protein serine/threonine phosphatase which is the protein phosphate 2A (PP2A). The PP2A involves the growth factor of the cell such as to induce the formation of mitogen-activated protein interaction[1] and playing a role in cell division and signal transduction. Therefore, PLM is used for the drug that prevents the tumor, cancer, or bacteria. There are nowsaday has 7 kinds of different PLM from PLM A to PLM G which differ the post-synthesis from the biosynthesis of PLM.
Phoslactomycin B (PLM B) is the product of the post synthase of the biosynthesis of phoslactomycin and the intermediate to produce the other PLMs. The biosynthesis of phoslactomycin belongs to type I polyketide synthase (PKS). A polyketide is are characterized by a macrocyclic lactone and is produced by bacteria and fungi. From the PLM B, there are many articles wrote about the synthesis of different PLM A through PLM G.
 | |
|---|---|
| Chemical and physical data | |
| Formula | |
| Vapour pressure[2] | 0.0±5.7 mmHg at 25 °C |
| Polarizability[2] | 54.3±0.5 10−24cm3 |
| Biology Organism | |
| Kingdom | Bacteria |
| Family | Streptomycetaceae |
| Identify Information | |
| Classification | OXIDOREDUCTASE |
|
C060449 |
| KNApSAcK CID | C00017683 |
| ChemBL | PMID 23790488 |
| Research PubMed | PMC3875705 |
lyketide synthase domains
The domains in the polyketide synthase type I:[3]
- ACP: acyl carrier protein (serves as chaperone)
- AT: acyl transferase (transfers acyl group form CoA to ACP)
- KS: keto synthase (forms the new carbon-carbon bond)
- KR: keto reductase (NADPH-dependent reduces beta-ketone to beta-hydroxyl)
- DH: dehydratase (eliminates beta-OH to anpha/beta-unsaturation)
- ER: enoyl reductase (NADPH-dependent reduces anpha/beta-unsaturation)
- TE: thioesterase (hydrolyses / cyclizes)
Biosynthesis pathway of phoslactomycin
The PKS of phoslactomycin has one loading domain, 7 modules and 6 proteins that encode PnA, PnB, PnC, PnD, PnE, and PnF. The biosynthesis starts the loading with the cyclohexyl- CoA. Stepping in each module, there always need the keto synthase (KS) to create the new linkage of carbon-carbon to elongate the chain, the acyl transferase to transfer acyl to ACP domain. Then ACP serves as the acyl carrier protein to the further reaction, and each module has the keto reductase at the end to reduce the ketone to hydroxyl group with more stable. Module 1 uses the precursor malonyl-CoA and dehydrase domain to create the double bond.
Similarly, module 2, module 5, and module 7 have the same 5 domains KS-AT-ACP-DH-KR, but module 7 has one more domain at the end is thioesterase (TE) to create the ring member of the phoslactomycin product. Module 4 and module 6 have 4 domains which are KS-AT-ACP-KR and use the precursor ethylmalonyl-CoA. The final product is phoslactomycin.
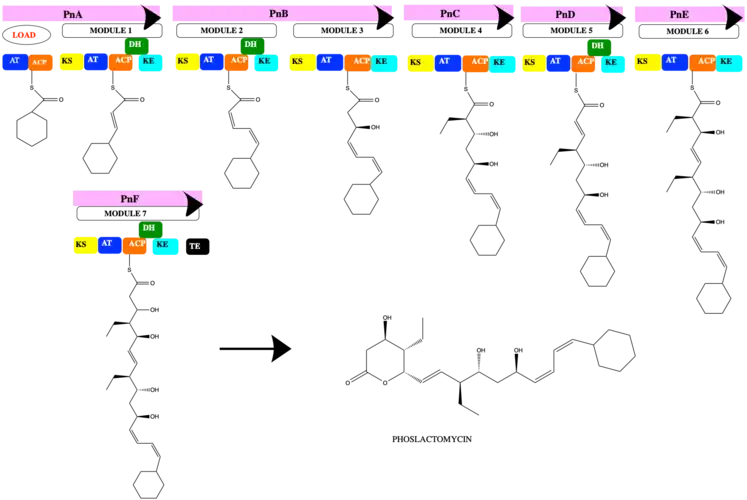
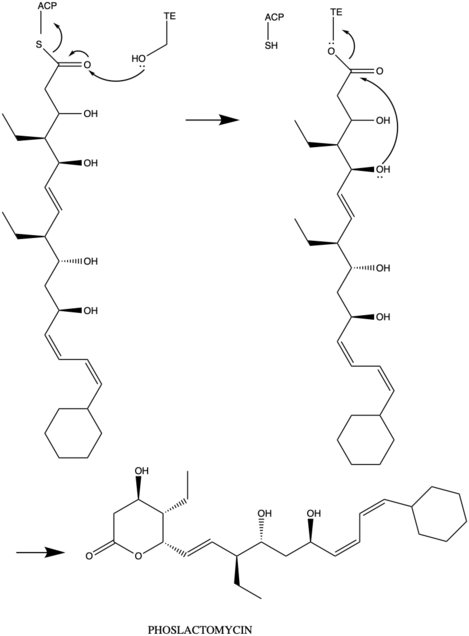
Phoslactomycin family
Isolation from Streptomyces platensis, PLM is produces. Genes PnT1 and PnT2 regulate the post-synthesis of PLM to form PLM B by the phosphorylation and added the amine group . Figure 4 is based on the biosynthesis analysis on the Gene journal introduced that PLM B is used to produce PLM A, and 4 more PLMs C-F.[4] The PLM A-F are the post synthesis product of the biosynthesis of PLM with the modification of many enzyme PnT1-T8.
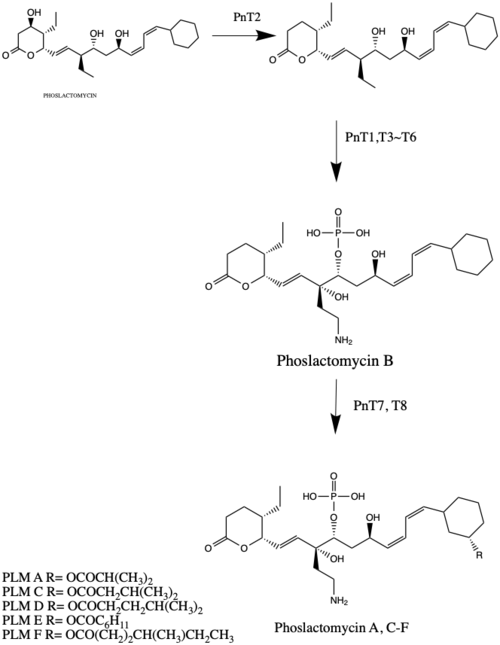
PLMs regulates the actin cytoskeleton as they induce actin depolymerization by the indirect way. In the experiment, PLM F does not affect to the polymerization of purified actin in vitro.[5] However, PLM F enhances the phosphorylation of intracellular vimentin.
Footnotes
- ↑ Klaasen and Watkins II. (2015). Casarett & Doull's Essentials of Tocicology (Third ed.). McGraw Hill, Lange. p. 29. ISBN 978-0-07-184709-4.
- 1 2 "Phoslactomycin B | C25H40NO8P | ChemSpider". www.chemspider.com. Retrieved 2020-05-18.
- ↑ Dewick, Paul M. (2009). Medicinal Natural Products (Third ed.). John Wiley & Sons. p. 39. ISBN 978-0-470-74168-9.
- ↑ Chen, Yun-Liang; Zhao, Juan; Liu, Wei; Gao, Ju-Fang; Tao, Li-Ming; Pan, Hai-Xue; Tang, Gong-Li (2012-11-10). "Identification of phoslactomycin biosynthetic gene clusters from Streptomyces platensis SAM-0654 and characterization of PnR1 and PnR2 as positive transcriptional regulators". Gene. 509 (2): 195–200. doi:10.1016/j.gene.2012.08.030. ISSN 0378-1119. PMID 22940146.
- ↑ Usui, Takeo; Marriott, Gerard; Inagaki, Masaki; Swarup, Ghanshyam; Osada, Hiroyuki (1999-05-01). "Protein Phosphatase 2A Inhibitors, Phoslactomycins. Effects on the Cytoskeleton in NIH/3T3 Cells". The Journal of Biochemistry. 125 (5): 960–965. doi:10.1093/oxfordjournals.jbchem.a022375. ISSN 0021-924X. PMID 10220590.