Photosystems are functional and structural units of protein complexes involved in photosynthesis. Together they carry out the primary photochemistry of photosynthesis: the absorption of light and the transfer of energy and electrons. Photosystems are found in the thylakoid membranes of plants, algae, and cyanobacteria. These membranes are located inside the chloroplasts of plants and algae, and in the cytoplasmic membrane of photosynthetic bacteria. There are two kinds of photosystems: PSI and PSII.
PSII will absorb red light, and PSI will absorb far-red light. Although photosynthetic activity will be detected when the photosystems are exposed to either red or far-red light, the photosynthetic activity will be the greatest when plants are exposed to both wavelengths of light. Studies have actually demonstrated that the two wavelengths together have a synergistic effect on the photosynthetic activity, rather than an additive one.[1]
Each photosystem has two parts: a reaction center, where the photochemistry occurs, and an antenna complex, which surrounds the reaction center. The antenna complex contains hundreds of chlorophyll molecules which funnel the excitation energy to the center of the photosystem. At the reaction center, the energy will be trapped and transferred to produce a high energy molecule.[2]
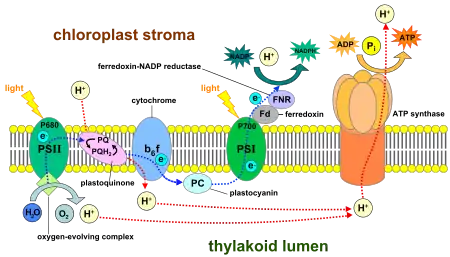
The main function of PSII is to efficiently split water into oxygen molecules and protons. PSII will provide a steady stream of electrons to PSI, which will boost these in energy and transfer them to NADP+ and H+ to make NADPH. The hydrogen from this NADPH can then be used in a number of different processes within the plant.[2]
Reaction centers
Reaction centers are multi-protein complexes found within the thylakoid membrane.
At the heart of a photosystem lies the reaction center, which is an enzyme that uses light to reduce and oxidize molecules (give off and take up electrons). This reaction center is surrounded by light-harvesting complexes that enhance the absorption of light.
In addition, surrounding the reaction center are pigments which will absorb light. The pigments which absorb light at the highest energy level are found furthest from the reaction center. On the other hand, the pigments with the lowest energy level are more closely associated with the reaction center. Energy will be efficiently transferred from the outer part of the antenna complex to the inner part. This funneling of energy is performed via resonance transfer, which occurs when energy from an excited molecule is transferred to a molecule in the ground state. This ground state molecule will be excited, and the process will continue between molecules all the way to the reaction center. At the reaction center, the electrons on the special chlorophyll molecule will be excited and ultimately transferred away by electron carriers. (If the electrons were not transferred away after excitation to a high energy state, they would lose energy by fluorescence back to the ground state, which would not allow plants to drive photosynthesis.) The reaction center will drive photosynthesis by taking light and turning it into chemical energy[3] that can then be used by the chloroplast.[2]
Two families of reaction centers in photosystems can be distinguished: type I reaction centers (such as photosystem I (P700) in chloroplasts and in green-sulfur bacteria) and type II reaction centers (such as photosystem II (P680) in chloroplasts and in non-sulfur purple bacteria). The two photosystems originated from a common ancestor, but have since diversified.[4][5]
Each of the photosystem can be identified by the wavelength of light to which it is most reactive (700 nanometers for PSI and 680 nanometers for PSII in chloroplasts), the amount and type of light-harvesting complex present, and the type of terminal electron acceptor used.
Type I photosystems use ferredoxin-like iron-sulfur cluster proteins as terminal electron acceptors, while type II photosystems ultimately shuttle electrons to a quinone terminal electron acceptor. Both reaction center types are present in chloroplasts and cyanobacteria, and work together to form a unique photosynthetic chain able to extract electrons from water, creating oxygen as a byproduct.
Structure of PSI and PSII
A reaction center comprises several (about 25-30)[6] protein subunits, which provide a scaffold for a series of cofactors. The cofactors can be pigments (like chlorophyll, pheophytin, carotenoids), quinones, or iron-sulfur clusters.[7]
Each photosystem has two main subunits: an antenna complex (a light harvesting complex or LHC) and a reaction center. The antenna complex is where light is captured, while the reaction center is where this light energy is transformed into chemical energy. At the reaction center, there are many polypeptides that are surrounded by pigment proteins. At the center of the reaction center is a special pair of chlorophyll molecules.
Each PSII has about 8 LHCII. These contain about 14 chlorophyll a and chlorophyll b molecules, as well as about four carotenoids. In the reaction center of PSII of plants and cyanobacteria, the light energy is used to split water into oxygen, protons, and electrons. The protons will be used in proton pumping to fuel the ATP synthase at the end of an electron transport chain. A majority of the reactions occur at the D1 and D2 subunits of PSII.
In oxygenic photosynthesis
Both photosystem I and II are required for oxygenic photosynthesis. Oxygenic photosynthesis can be performed by plants and cyanobacteria; cyanobacteria are believed to be the progenitors of the photosystem-containing chloroplasts of eukaryotes. Photosynthetic bacteria that cannot produce oxygen have only one photosystem, which is similar to either PSI or PSII.
At the core of photosystem II is P680, a special chlorophyll to which incoming excitation energy from the antenna complex is funneled. One of the electrons of excited P680* will be transferred to a non-fluorescent molecule, which ionizes the chlorophyll and boosts its energy further, enough that it can split water in the oxygen evolving complex (OEC) of PSI and recover its electron. At the heart of the OEC are 4 Mn atoms, each of which can trap one electron. The electrons harvested from the splitting of two waters fill the OEC complex in its highest-energy state, which holds 4 excess electrons.[2]
Electrons travel through the cytochrome b6f complex to photosystem I via an electron transport chain within the thylakoid membrane. Energy from PSI drives this process and is harnessed (the whole process is termed chemiosmosis) to pump protons across the membrane, into the thylakoid lumen space from the chloroplast stroma. This will provide a potential energy difference between lumen and stroma, which amounts to a proton-motive force that can be utilized by the proton-driven ATP synthase to generate ATP. If electrons only pass through once, the process is termed noncyclic photophosphorylation, but if they pass through PSI and the proton pump multiple times it is called cyclic photophosphorylation.
When the electron reaches photosystem I, it fills the electron deficit of light-excited reaction-center chlorophyll P700+ of PSI. The electron may either continue to go through cyclic electron transport around PSI or pass, via ferredoxin, to the enzyme NADP+ reductase. Electrons and protons are added to NADP+ to form NADPH. This reducing (hydrogenation) agent is transported to the Calvin cycle to react with glycerate 3-phosphate, along with ATP to form glyceraldehyde 3-phosphate, the basic building block from which plants can make a variety of substances.
Photosystem repair
In intense light, plants use various mechanisms to prevent damage to their photosystems. They are able to release some light energy as heat, but the excess light can also produce reactive oxygen species. While some of these can be detoxified by antioxidants, the remaining oxygen species will be detrimental to the photosystems of the plant. More specifically, the D1 subunit in the reaction center of PSII can be damaged. Studies have found that deg1 proteins are involved in the degradation of these damaged D1 subunits. New D1 subunits can then replace these damaged D1 subunits in order to allow PSII to function properly again.[8]
See also
References
- ↑ Zhen, Shuyang; Van Iersel, Marc W. (2017-02-01). "Far-red light is needed for efficient photochemistry and photosynthesis". Journal of Plant Physiology. 209: 115–122. doi:10.1016/j.jplph.2016.12.004. ISSN 0176-1617. PMID 28039776.
- 1 2 3 4 Taiz, Lincoln (2018). Fundamentals of plant physiology. ISBN 978-1-60535-790-4. OCLC 1035316853.
- ↑ Gisriel, Christopher; Sarrou, Iosifina; Ferlez, Bryan; Golbeck, John H.; Redding, Kevin E.; Fromme, Raimund (2017-09-08). "Structure of a symmetric photosynthetic reaction center–photosystem". Science. 357 (6355): 1021–1025. Bibcode:2017Sci...357.1021G. doi:10.1126/science.aan5611. ISSN 0036-8075. PMID 28751471.
- ↑ Sadekar S, Raymond J, Blankenship RE (November 2006). "Conservation of distantly related membrane proteins: photosynthetic reaction centers share a common structural core". Molecular Biology and Evolution. 23 (11): 2001–7. doi:10.1093/molbev/msl079. PMID 16887904.
- ↑ Orf GS, Gisriel C, Redding KE (October 2018). "Evolution of photosynthetic reaction centers: insights from the structure of the heliobacterial reaction center". Photosynthesis Research. 138 (1): 11–37. doi:10.1007/s11120-018-0503-2. OSTI 1494566. PMID 29603081. S2CID 4473759.
- ↑ Caffarri, Stefano; Tibiletti, Tania; Jennings, Robert C.; Santabarbara, Stefano (June 2014). "A Comparison Between Plant Photosystem I and Photosystem II Architecture and Functioning". Current Protein & Peptide Science. 15 (4): 296–331. doi:10.2174/1389203715666140327102218. ISSN 1389-2037. PMC 4030627. PMID 24678674.
- ↑ Jagannathan, B; Golbeck, JH (2009). Photosynthesis:Microbial. pp. 325–341. doi:10.1016/B978-012373944-5.00352-7. ISBN 9780123739445.
{{cite book}}:|journal=ignored (help) - ↑ Kapri-Pardes, Einat; Naveh, Leah; Adam, Zach (March 2007). "The Thylakoid Lumen Protease Deg1 Is Involved in the Repair of Photosystem II from Photoinhibition in Arabidopsis". The Plant Cell. 19 (3): 1039–1047. doi:10.1105/tpc.106.046573. ISSN 1040-4651. PMC 1867356. PMID 17351117.
External links
- Photosystems I + II: Imperial College, Barber Group
- Photosystem I: Molecule of the Month in the Protein Data Bank
- Photosystem II: Molecule of the Month in the Protein Data Bank
- Photosystem II: ANU
- UMich Orientation of Proteins in Membranes Superfamily » 1.1.002. Photosystems (7 families) - Orientations of Proteins in Membranes (OPM) database at the Wayback Machine (archived October 13, 2017) – Calculated spatial positions of photosynthetic reaction centers and photosystems in membrane
- Rutherford AW, Faller P (January 2003). "Photosystem II: evolutionary perspectives". Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 (1429): 245–53. doi:10.1098/rstb.2002.1186. PMC 1693113. PMID 12594932.