| preQ1-II (pre queuosine) riboswitch | |
|---|---|
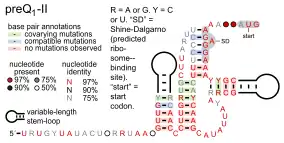
 Predicted secondary structure and sequence conservation of preQ1-II | |
| Identifiers | |
| Symbol | preQ1-II |
| Alt. Symbols | COG4708 |
| Rfam | RF01054 |
| Other data | |
| RNA type | Cis-reg; riboswitch |
| Domain(s) | Bacteria |
| SO | SO:0005836 |
| PDB structures | PDBe |
PreQ1-II riboswitches form a class of riboswitches that specifically bind pre-queuosine1 (PreQ1), a precursor of the modified nucleoside queuosine.[1] They are found in certain species of Streptococcus and Lactococcus, and were originally identified as a conserved RNA secondary structure called the "COG4708 motif".[2] All known members of this riboswitch class appear to control members of COG4708 genes. These genes are predicted to encode membrane-bound proteins and have been proposed to be a transporter of preQ1, or a related metabolite, based on their association with preQ1-binding riboswitches.[1] PreQ1-II riboswitches have no apparent similarities in sequence or structure to preQ1-I riboswitches, a previously discovered class of preQ1-binding riboswitches. PreQ1 thus joins S-adenosylmethionine as the second metabolite to be found that is the ligand of more than one riboswitch class.
References
- 1 2 3 Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR (2008). "Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria". RNA. 14 (4): 685–695. doi:10.1261/rna.937308. PMC 2271366. PMID 18305186.
- 1 2 Weinberg Z, Barrick JE, Yao Z, et al. (2007). "Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline". Nucleic Acids Res. 35 (14): 4809–4819. doi:10.1093/nar/gkm487. PMC 1950547. PMID 17621584.