| ROOL | |
|---|---|
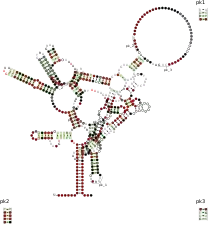 Consensus secondary structure and sequence conservation of ROOL RNA | |
| Identifiers | |
| Symbol | ROOL |
| Rfam | RF03087 |
| Other data | |
| RNA type | Gene; sRNA |
| SO | SO:0001263 |
| PDB structures | PDBe |
The Rumen-Originating, Ornate, Large (ROOL) RNA motif was originally discovered by bioinformatics by analyzing metagenomic sequences from cow rumen.[1] ROOL RNAs are found in a variety of bacterial species and apparently do not code for proteins. The RNA has a complex RNA secondary structure and its average size of 581 nucleotides[1] is unusually large for bacterial non-coding RNAs. This large size and structural complexity for a bacterial RNA is consistent with properties of large ribozymes.[2]
ROOL RNAs are present in prophages and purified phages, but also in bacterial genomic locations that do not appear to be related to phages.[1] ROOL RNAs are also frequently located nearby to tRNAs. The large size and complicated secondary structure of ROOL RNAs, combined with their association with tRNAs and phages are properties that are shared by the GOLLD RNA motif, another bacterial non-coding RNA. These shared properties could suggest a related function, but the commonalities could arise for other reasons.[1]
ROOL RNAs are present in bacteria in the phyla Bacillota, Fusobacteriota and Mycoplasmatota,[1] in addition to phages and cow rumen metagenomes, as mentioned above. Within the Bacillota, they are present in a few species of Clostridia and many lactic acid bacteria, especially those in the genus Lactobacillus.
ROOL RNAs in Lactobacillus salivarius were independently discovered based on their extremely high rates of expression in Lactobacillus salivarius UCC118[3] ROOL RNAs in various strains of Lactobacillus salivarius were then studied; there is a very large range in the expression levels of ROOL RNAs in different strains, and some strains do not appear to have these RNAs in their genome.[3] ROOL RNAs in L. salivarius UC118 are so abundant in some growth conditions that in late stationary phase they exceed even 16S ribosomal RNAs.[3] The experimentally determined size for ROOL RNAs in L. salivarius[3] closely matches the size of the proposed RNA structure based on bioinformatics.[1]
References
- 1 2 3 4 5 6 Weinberg, Zasha; Lünse, Christina E.; Corbino, Keith A.; Ames, Tyler D.; Nelson, James W.; Roth, Adam; Perkins, Kevin R.; Sherlock, Madeline E.; Breaker, Ronald R. (10 August 2017). "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Research. 45 (18): 10811–10823. doi:10.1093/nar/gkx699. PMC 5737381. PMID 28977401.
- ↑ Weinberg Z, Perreault J, Meyer MM, Breaker RR (December 2009). "Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis". Nature. 462 (7273): 656–659. Bibcode:2009Natur.462..656W. doi:10.1038/nature08586. PMC 4140389. PMID 19956260.
- 1 2 3 4 Cousin, Fabien J.; Lynch, Denise B.; Chuat, Victoria; Bourin, Maxence J. B.; Casey, Pat G.; Dalmasso, Marion; Harris, Hugh M. B.; McCann, Angela; O’Toole, Paul W. (17 July 2017). "A long and abundant non-coding RNA in Lactobacillus salivarius". Microbial Genomics. 3 (9): e000126. doi:10.1099/mgen.0.000126. PMC 5643018. PMID 29114404.