| Spiroplasma kunkelii | |
|---|---|
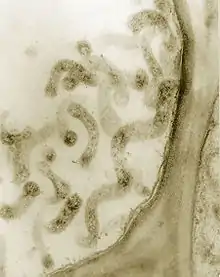 | |
| Corn stunt Spiroplasma in phloem cells. Thick section (0.4 micrometers) observed in a TEM. Magnified 75,000X. | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Mycoplasmatota |
| Class: | Mollicutes |
| Order: | Mycoplasmatales |
| Family: | Mycoplasmataceae |
| Genus: | Spiroplasma |
| Species: | S. kunkelii |
| Binomial name | |
| Spiroplasma kunkelii Whitcomb, Chen et al. | |
Spiroplasma kunkelii is a species of Mollicutes, which are small bacteria that all share a common cell wall-less feature.[1] They are characterized by helical and spherical morphology, they actually have the ability to be spherical or helical depending on the circumstances. The cells movement is bound by a membrane. The cell size ranges from 0.15 to 0.20 micrometers.[2]
Morphology
Spiroplasma kunkelii is a helical prokaryote that does not have a cell wall. The helical shape of S. kunkelii allows for the bacterium to be motile through flexional and rotational motility. The cell sizes are approximately 0.15-0.2 µm in diameter and 2.0 15 µm in length.[2] The elongated shape of S. kunkelii aids in nutrient import. The helical shape is thought to be the result of the cytoskeletal protein fibril. Though it has been observed that with environmental changes S. kunkelii can change into a coccoid shape. The ends of the helical shape have a tapered ends and blunt or rounded tips. The tip structure exposes adhesins that are used to attach to cells.[3]
Genome and phylogeny
Initial phylogeny of S. kunkelii was established according to biochemical and phenotypic characteristics. These methods provided a general phylogeny, but large gaps in knowledge were made apparent when considering the possibility of horizontal gene transfer. The genome has some unique identities, the chromosomal structure if circular and plasmids are currently not found on the chromosome. The chromosome CR2-3X is the major chromosome that has been focused on the most. There are 27 genes located on that chromosome, 23 of those genes are involved in chromosome partitioning, DNA replication, transcription, and translation. The small genomes of Spiroplasmas is close to the minimal complement necessary for life and pathogenesis. Their ability to survive in specific niches gives in sight to unique ability of microorganisms to be able to evolve pathogenesis while experiencing severe genome reductions. Due to the nature of bacteria and their abilities to transfer genetic material through horizontal gene transfer, phylogenies and genome histories are difficult to define. Studies mostly focus on the analysis of the 16S rRNA gene to define relations and build phylogenic trees. However, to confirm the connections made from 16S rRNA analysis studies have started to analyze the house keeping genes within S. kunkelii.[4]
Full analysis of the S. kunkelii genome have been done and the complete results have been posted to the GenBank. Studies have shown that Spiroplasma citri, Spiroplasma phoencium and Spiroplasma melliferum genomes bare the closest relations to S. kunkelii.[5]
Pathology
Spiroplasma kunkelii is an insect pathogen, it is transferred to other organisms by insects and it uses the host insect vector to multiply, but it is also important to note that some of the hosts are within the plants. Although they are transferred by insects, Spiroplasma kunkelii need plants in order to survive and grow. Spiroplasma kunkelii have a mutualistic relationship between arthropods and plants. Hosts include Zea mays, Zea mexicana and Zea perennis.[2]
Metabolism
Spiroplasma kunkelii have simple metabolism. They tend to grow well between 35-38 C and around the normal temperature of the human body. Spiroplasma kunkelii uses the host insect vector for multiplication, but it uses plants for survival.[6]
Little research has been done regarding the metabolism of S. kunkelii in detail. From the genetic relations many of the characteristics of their metabolism have been inferred from the Spiroplasma family and their close relative S. citri. Spiroplasma are chemoorganoheterotrophs due to their consumption of organic carbon and their parasitic lifestyle.[7] S. kunkelii utilizes the Embden-Meyerhof-Parnas pathway. Studies have found that S. kunkelii lack cytidine, dCMP deaminases, transaldolase and deoxyribose 5-phosphate aldolase enzymes.[8]
Corn stunt disease
The common name of Spiroplasma kunkelii is corn stunt spiroplasma, as it is known to cause corn stunt disease. It is considered a significant economic risk.[9] Corn stunt disease results in smaller corn husks and loose or missing kernels. The corn industry is a billion-dollar industry that is already being threatened by global warming and the introduction of S. kunkelii increases the loss possible in the industry.[10] S. kunkelii is spreading and has been found in south eastern United states. Though cases of S. kunkelii are take very seriously, due to it being a vector borne pathogen, it is able to be transferred between plants while only requiring one infected insect. Studies are on going to find proper resistance to this pathogen and will be progressing along with the disease.[11]
References
- ↑ "Spiroplasma--discovery and phylogeny". Agricultural Research Service (ARS). U.S. Department of Agriculture. Retrieved 5 December 2021.
- 1 2 3 "Spiroplasma kunkelii (corn stunt spiroplasma)". Invasive Species Compendium. Centre for Agriculture and Bioscience International (CABI). Retrieved 5 December 2021.
- ↑ Harne S, Gayathri P, Béven L (2020-10-21). "Exploring Spiroplasma Biology: Opportunities and Challenges". Frontiers in Microbiology. 11: 589279. doi:10.3389/fmicb.2020.589279. PMC 7609405. PMID 33193251.
- ↑ Zhao Y, Davis RE, Lee IM (September 2005). "Phylogenetic positions of 'Candidatus Phytoplasma asteris' and Spiroplasma kunkelii as inferred from multiple sets of concatenated core housekeeping proteins". International Journal of Systematic and Evolutionary Microbiology. 55 (Pt 5): 2131–2141. doi:10.1099/ijs.0.63655-0. PMID 16166721.
- ↑ Davis RE, Shao J, Dally EL, Zhao Y, Gasparich GE, Gaynor BJ, et al. (October 2015). "Complete Genome Sequence of Spiroplasma kunkelii Strain CR2-3x, Causal Agent of Corn Stunt Disease in Zea mays L". Genome Announcements. 3 (5): e01216–15. doi:10.1128/genomeA.01216-15. PMC 4616174. PMID 26494665.
- ↑ Bai X, Hogenhout SA (April 2002). "A genome sequence survey of the mollicute corn stunt spiroplasma Spiroplasma kunkelii". FEMS Microbiology Letters. 210 (1): 7–17. doi:10.1111/j.1574-6968.2002.tb11153.x. PMID 12023071.
- ↑ Renaudin J (2006). "Sugar metabolism and pathogenicity of Spiroplasma citri". Journal of Plant Pathology. 88 (2): 129–139. ISSN 1125-4653. JSTOR 41998303.
- ↑ Pollack JD, McElwain MC, DeSantis D, Manolukas JT, Tully JG, Chang CJ, et al. (October 1989). "Metabolism of members of the Spiroplasmataceae". International Journal of Systematic and Evolutionary Microbiology. 39 (4): 406–12. doi:10.1099/00207713-39-4-406.
- ↑ Carroll MW, Glaser JA, Hellmich RL, Hunt TE, Sappington TW, Calvin D, Copenhaver K, Fridgen J (October 2008). "Use of spectral vegetation indices derived from airborne hyperspectral imagery for detection of European corn borer infestation in Iowa corn plots". Journal of Economic Entomology. 101 (5): 1614–23. doi:10.1603/0022-0493(2008)101[1614:uosvid]2.0.co;2. PMID 18950044.
- ↑ Chavas JP, Kim K, Lauer JG, Klemme RM, Bland WL (2001). "An Economic Analysis of Corn Yield, Corn Profitability, and Risk at the Edge of the Corn Belt". Journal of Agricultural and Resource Economics. 26 (1): 230–247. ISSN 1068-5502. JSTOR 40987105.
- ↑ Oleszczuk JD, Catalano MI, Dalaisón L, Di Rienzo JA, Giménez Pecci MP, Carpane P (2020). "Characterization of components of resistance to Corn Stunt disease". PLOS ONE. 15 (10): e0234454. Bibcode:2020PLoSO..1534454O. doi:10.1371/journal.pone.0234454. PMC 7571704. PMID 33075073.