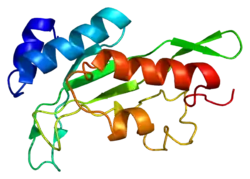
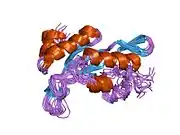
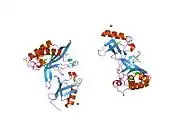
Tumor susceptibility gene 101, also known as TSG101, is a human gene that encodes for a cellular protein of the same name.
Function
The protein encoded by this gene belongs to a group of apparently inactive homologs of ubiquitin-conjugating enzymes. The gene product contains a coiled-coil domain that interacts with stathmin, a cytosolic phosphoprotein implicated in tumorigenesis. The protein may play a role in cell growth and differentiation and act as a negative growth regulator. In vitro steady-state expression of this tumor susceptibility gene appears to be important for maintenance of genomic stability and cell cycle regulation. Mutations and alternative splicing in this gene occur in high frequency in breast cancer and suggest that defects occur during breast cancer tumorigenesis and/or progression.[5]
The main role of TSG101 is to participate in ESCRT pathway. This pathway facilitates reverse topology budding and formation of multivesicular bodies (MVB) which delivers cargo destined for degradation to the lysosomes.[6] TSG101 recognises short linear motif : P(T/S)AP via the UEV protein domain of the VPS23/TSG101 subunit. The assembly of the ESCRT-I complex is directed by the C-terminal steadiness box (SB) of VPS23, the N-terminal half of VPS28, and the C-terminal half of VPS37. The structure is primarily composed of three long, parallel helical hairpins, each corresponding to a different subunit. The additional domains and motifs extending beyond the core serve as gripping tools for ESCRT-I critical functions.[7][8]
Viral Hijacking
TSG101 plays an important role in the pathogenesis of HIV and other viruses. In uninfected cells, TSG101 functions in the biogenesis of the multivesicular body (MVB),[9] which suggests that HIV may bind TSG101 in order to gain access to the downstream machinery that catalyzes MVB vesicle budding.[10]
Interactions
TSG101 has been shown to interact with:
Orthologue, Vps23
| Vps23_core | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 escrt-i core | |||||||||
| Identifiers | |||||||||
| Symbol | Vps23_core | ||||||||
| Pfam | PF09454 | ||||||||
| InterPro | IPR017916 | ||||||||
| |||||||||
In humans, the orthologue of vps23 which has a component of ESCRT-1 is called Tsg101. Mutations in Tsg-101 have been linked to cervical, breast, prostate and gastrointestinal cancers. In molecular biology, vps23 (vacuolar protein sorting) is a protein domain. Vps proteins are components of the ESCRTs (endosomal sorting complexes required for transport) which are required for protein sorting at the early endosome. More specifically, vps23 is a component of ESCRT-I. The ESCRT complexes form the machinery driving protein sorting from endosomes to lysosomes. ESCRT complexes are central to receptor down-regulation, lysosome biogenesis and budding of HIV.
Structure
Yeast ESCRT-I consists of three protein subunits, VPS23, VPS28, and VPS37. In humans, ESCRT-I comprises TSG101, VPS28, and one of four potential human VPS37 homologues.
See also
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000074319 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000014402 - Ensembl, May 2017
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Entrez Gene: TSG101 tumor susceptibility gene 101".
- ↑ Henne WM, Buchkovich NJ, Emr SD (July 2011). "The ESCRT pathway". Developmental Cell. 21 (1): 77–91. doi:10.1016/j.devcel.2011.05.015. PMID 21763610.
- ↑ Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL (April 2006). "ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes". Cell. 125 (1): 99–111. doi:10.1016/j.cell.2006.01.047. PMID 16615893. S2CID 14017291.
- ↑ Kostelansky MS, Sun J, Lee S, Kim J, Ghirlando R, Hierro A, Emr SD, Hurley JH (April 2006). "Structural and functional organization of the ESCRT-I trafficking complex". Cell. 125 (1): 113–26. doi:10.1016/j.cell.2006.01.049. PMC 1576341. PMID 16615894.
- ↑ Katzmann DJ, Odorizzi G, Emr SD (2002). "Receptor downregulation and multivesicular-body sorting". Nat. Rev. Mol. Cell Biol. 3 (12): 893–905. doi:10.1038/nrm973. PMID 12461556. S2CID 1344520.
- ↑ von Schwedler UK, Stuchell M, Müller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Kräusslich HG, Kaplan J, Morham SG, Sundquist WI (2003). "The protein network of HIV budding". Cell. 114 (6): 701–13. doi:10.1016/S0092-8674(03)00714-1. PMID 14505570. S2CID 16894972.
- ↑ Sun Z, Pan J, Hope WX, Cohen SN, Balk SP (August 1999). "Tumor susceptibility gene 101 protein represses androgen receptor transactivation and interacts with p300". Cancer. 86 (4): 689–96. doi:10.1002/(sici)1097-0142(19990815)86:4<689::aid-cncr19>3.0.co;2-p. PMID 10440698. S2CID 26971556.
- 1 2 3 Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- ↑ Lu Q, Hope LW, Brasch M, Reinhard C, Cohen SN (June 2003). "TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation". Proc. Natl. Acad. Sci. U.S.A. 100 (13): 7626–31. Bibcode:2003PNAS..100.7626L. doi:10.1073/pnas.0932599100. PMC 164637. PMID 12802020.
- ↑ Amit I, Yakir L, Katz M, Zwang Y, Marmor MD, Citri A, Shtiegman K, Alroy I, Tuvia S, Reiss Y, Roubini E, Cohen M, Wides R, Bacharach E, Schubert U, Yarden Y (July 2004). "Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding". Genes Dev. 18 (14): 1737–52. doi:10.1101/gad.294904. PMC 478194. PMID 15256501.
- ↑ Oh H, Mammucari C, Nenci A, Cabodi S, Cohen SN, Dotto GP (April 2002). "Negative regulation of cell growth and differentiation by TSG101 through association with p21(Cip1/WAF1)". Proc. Natl. Acad. Sci. U.S.A. 99 (8): 5430–5. Bibcode:2002PNAS...99.5430O. doi:10.1073/pnas.082123999. PMC 122786. PMID 11943869.
- ↑ Li L, Liao J, Ruland J, Mak TW, Cohen SN (February 2001). "A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control". Proc. Natl. Acad. Sci. U.S.A. 98 (4): 1619–24. Bibcode:2001PNAS...98.1619L. doi:10.1073/pnas.98.4.1619. PMC 29306. PMID 11172000.
- ↑ Stuchell MD, Garrus JE, Müller B, Stray KM, Ghaffarian S, McKinnon R, Kräusslich HG, Morham SG, Sundquist WI (August 2004). "The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding". J. Biol. Chem. 279 (34): 36059–71. doi:10.1074/jbc.M405226200. PMID 15218037.
- ↑ Bishop N, Woodman P (April 2001). "TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes". J. Biol. Chem. 276 (15): 11735–42. doi:10.1074/jbc.M009863200. PMID 11134028.
Further reading
- Mazzé FM, Degrève L (2006). "The role of viral and cellular proteins in the budding of human immunodeficiency virus". Acta Virol. 50 (2): 75–85. PMID 16808324.
- Freed EO, Mouland AJ (2006). "The cell biology of HIV-1 and other retroviruses". Retrovirology. 3: 77. doi:10.1186/1742-4690-3-77. PMC 1635732. PMID 17083721.
- Li L, Li X, Francke U, Cohen SN (1997). "The TSG101 tumor susceptibility gene is located in chromosome 11 band p15 and is mutated in human breast cancer". Cell. 88 (1): 143–54. doi:10.1016/S0092-8674(00)81866-8. PMID 9019400.
- Koonin EV, Abagyan RA (1997). "TSG101 may be the prototype of a class of dominant negative ubiquitin regulators". Nat. Genet. 16 (4): 330–1. doi:10.1038/ng0897-330. PMID 9241264. S2CID 36275138.
- Steiner P, Barnes DM, Harris WH, Weinberg RA (1997). "Absence of rearrangements in the tumour susceptibility gene TSG101 in human breast cancer". Nat. Genet. 16 (4): 332–3. doi:10.1038/ng0897-332. PMID 9241265. S2CID 20233125.
- Lee MP, Feinberg AP (1997). "Aberrant splicing but not mutations of TSG101 in human breast cancer". Cancer Res. 57 (15): 3131–4. PMID 9242438.
- Gayther SA, Barski P, Batley SJ, Li L, de Foy KA, Cohen SN, Ponder BA, Caldas C (1997). "Aberrant splicing of the TSG101 and FHIT genes occurs frequently in multiple malignancies and in normal tissues and mimics alterations previously described in tumours". Oncogene. 15 (17): 2119–26. doi:10.1038/sj.onc.1201591. PMID 9366528. S2CID 7819803.
- Xie W, Li L, Cohen SN (1998). "Cell cycle-dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency". Proc. Natl. Acad. Sci. U.S.A. 95 (4): 1595–600. Bibcode:1998PNAS...95.1595X. doi:10.1073/pnas.95.4.1595. PMC 19109. PMID 9465061.
- Wagner KU, Dierisseau P, Rucker EB, Robinson GW, Hennighausen L (1998). "Genomic architecture and transcriptional activation of the mouse and human tumor susceptibility gene TSG101: common types of shorter transcripts are true alternative splice variants". Oncogene. 17 (21): 2761–70. doi:10.1038/sj.onc.1202529. PMID 9840940. S2CID 29505462.
- Sun Z, Pan J, Hope WX, Cohen SN, Balk SP (1999). "Tumor susceptibility gene 101 protein represses androgen receptor transactivation and interacts with p300". Cancer. 86 (4): 689–96. doi:10.1002/(SICI)1097-0142(19990815)86:4<689::AID-CNCR19>3.0.CO;2-P. PMID 10440698. S2CID 26971556.
- Hittelman AB, Burakov D, Iñiguez-Lluhí JA, Freedman LP, Garabedian MJ (1999). "Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins". EMBO J. 18 (19): 5380–8. doi:10.1093/emboj/18.19.5380. PMC 1171607. PMID 10508170.
- Rountree MR, Bachman KE, Baylin SB (2000). "DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci". Nat. Genet. 25 (3): 269–77. doi:10.1038/77023. PMID 10888872. S2CID 26149386.
- Bishop N, Woodman P (2001). "TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes". J. Biol. Chem. 276 (15): 11735–42. doi:10.1074/jbc.M009863200. PMID 11134028.
- Li L, Liao J, Ruland J, Mak TW, Cohen SN (2001). "A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control". Proc. Natl. Acad. Sci. U.S.A. 98 (4): 1619–24. Bibcode:2001PNAS...98.1619L. doi:10.1073/pnas.98.4.1619. PMC 29306. PMID 11172000.
- VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA (2001). "Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag)". Proc. Natl. Acad. Sci. U.S.A. 98 (14): 7724–9. Bibcode:2001PNAS...98.7724V. doi:10.1073/pnas.131059198. PMC 35409. PMID 11427703.
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL, Myszka DG, Sundquist WI (2001). "Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding". Cell. 107 (1): 55–65. doi:10.1016/S0092-8674(01)00506-2. PMID 11595185. S2CID 18525780.
- Martin-Serrano J, Zang T, Bieniasz PD (2002). "HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress". Nat. Med. 7 (12): 1313–9. doi:10.1038/nm1201-1313. PMID 11726971. S2CID 19655035.
- Demirov DG, Ono A, Orenstein JM, Freed EO (2002). "Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function". Proc. Natl. Acad. Sci. U.S.A. 99 (2): 955–60. Bibcode:2002PNAS...99..955D. doi:10.1073/pnas.032511899. PMC 117412. PMID 11805336.
- Bennett NA, Pattillo RA, Lin RS, Hsieh CY, Murphy T, Lyn D (2002). "TSG101 expression in gynecological tumors: relationship to cyclin D1, cyclin E, p53 and p16 proteins". Cell. Mol. Biol. (Noisy-le-grand). 47 (7): 1187–93. PMID 11838966.
- Bishop N, Horman A, Woodman P (2002). "Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates". J. Cell Biol. 157 (1): 91–101. doi:10.1083/jcb.200112080. PMC 2173266. PMID 11916981.