Trophic coherence is a property of directed graphs (or directed networks).[1] It is based on the concept of trophic levels used mainly in ecology,[2] but which can be defined for directed networks in general and provides a measure of hierarchical structure among nodes. Trophic coherence is the tendency of nodes to fall into well-defined trophic levels. It has been related to several structural and dynamical properties of directed networks, including the prevalence of cycles[3] and network motifs,[4] ecological stability,[1] intervality,[5] and spreading processes like epidemics and neuronal avalanches.[6]
Definition
Consider a directed network defined by the adjacency matrix . Each node can be assigned a trophic level according to
where is 's in-degree, and nodes with (basal nodes) have by convention. Each edge has a trophic difference associated, defined as . The trophic coherence of the network is a measure of how tightly peaked the distribution of trophic distances, , is around its mean value, which is always . This can be captured by an incoherence parameter , equal to the standard deviation of :
where is the number of edges in the network.[1]
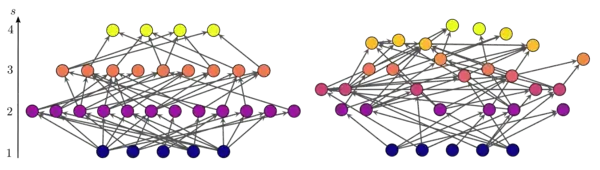
The figure shows two networks which differ in their trophic coherence. The position of the nodes on the vertical axis corresponds to their trophic level. In the network on the left, nodes fall into distinct (integer) trophic levels, so the network is maximally coherent . In the one on the right, many of the nodes have fractional trophic levels, and the network is more incoherent .[6]
Trophic coherence in nature
The degree to which empirical networks are trophically coherent (or incoherent) can be investigated by comparison with a null model. This is provided by the basal ensemble, which comprises networks in which all non-basal nodes have the same proportion of basal nodes for in-neighbours.[3] Expected values in this ensemble converge to those of the widely used configuration ensemble[7] in the limit , (with and the numbers of nodes and edges), and can be shown numerically to be a good approximation for finite random networks. The basal ensemble expectation for the incoherence parameter is
where is the number of edges connected to basal nodes.[3] The ratio measured in empirical networks reveals whether they are more or less coherent than the random expectation. For instance, Johnson and Jones[3] find in a set of networks that food webs are significantly coherent , metabolic networks are significantly incoherent , and gene regulatory networks are close to the random expectation .
Trophic levels and node function
There is as yet little understanding of the mechanisms which might lead to particular kinds of networks becoming significantly coherent or incoherent.[3] However, in systems which present correlations between trophic level and other features of nodes, processes which tended to favour the creation of edges between nodes with particular characteristics could induce coherence or incoherence. In the case of food webs, predators tend to specialise on consuming prey with certain biological properties (such as size, speed or behaviour) which correlate with their diet, and hence with trophic level. This has been suggested as the reason for food-web coherence.[1] However, food-web models based on a niche axis do not reproduce realistic trophic coherence,[1] which may mean either that this explanation is insufficient, or that several niche dimensions need to be considered.[8]

The relation between trophic level and node function can be seen in networks other than food webs. The figure shows a word adjacency network derived from the book Green Eggs and Ham, by Dr. Seuss.[3] The height of nodes represents their trophic levels (according here to the edge direction which is the opposite of that suggested by the arrows, which indicate the order in which words are concatenated in sentences). The syntactic function of words is also shown with node colour. There is a clear relationship between syntactic function and trophic level: the mean trophic level of common nouns (blue) is , whereas that of verbs (red) is . This example illustrates how trophic coherence or incoherence might emerge from node function, and also that the trophic structure of networks provides a means of identifying node function in certain systems.
Generating trophically coherent networks
There are various ways of generating directed networks with specified trophic coherence, all based on gradually introducing new edges to the system in such a way that the probability of each new candidate edge being accepted depends on the expected trophic difference it would have.
The preferential preying model is an evolving network model similar to the Barábasi-Albert model of preferential attachment, but inspired on an ecosystem that grows through immigration of new species.[1] One begins with basal nodes and proceeds to introduce new nodes up to a total of . Each new node is assigned a first in-neighbour (a prey species in the food-web context) and a new edge is placed from to . The new node is given a temporary trophic level . Then a further new in-neighbours are chosen for from among those in the network according to their trophic levels. Specifically, for a new candidate in-neighbour , the probability of being chosen is a function of . Johnson et al[1] use
where is a parameter which tunes the trophic coherence: for maximally coherent networks are generated, and increases monotonically with for . The choice of is arbitrary. One possibility is to set to , where is the number of nodes already in the network when arrives, and is a random variable drawn from a Beta distribution with parameters and
( being the desired number of edges). This way, the generalised cascade model[9][10] is recovered in the limit , and the degree distributions are as in the niche model[11] and generalised niche model.[10] This algorithm, as described above, generates networks with no cycles (except for self-cycles, if the new node is itself considered among its candidate in-neighbours ). In order for cycles of all lengths to be a possible, one can consider new candidate edges in which the new node is the in-neighbour as well as those in which it would be the out-neighbour. The probability of acceptance of these edges, , then depends on .
The generalised preferential preying model[6] is similar to the one described above, but has certain advantages. In particular, it is more analytically tractable, and one can generate networks with a precise number of edges . The network begins with basal nodes, and then a further new nodes are added in the following way. When each enters the system, it is assigned a single in-neighbour randomly from among those already there. Every node then has an integer temporary trophic level . The remaining edges are introduced as follows. Each pair of nodes has two temporary trophic distances associated, and . Each of these candidate edges is accepted with a probability that depends on this temporary distance. Klaise and Johnson[6] use
because they find the distribution of trophic distances in several kinds of networks to be approximately normal, and this choice leads to a range of the parameter in which . Once all the edges have been introduced, one must recalculate the trophic levels of all nodes, since these will differ from the temporary ones originally assigned unless . As with the preferential preying model, the average incoherence parameter of the resulting networks is a monotonically increasing function of for . The figure above shows two networks with different trophic coherence generated with this algorithm.
References
- 1 2 3 4 5 6 7 Johnson S, Domı́nguez-Garcı́a V, Donetti L, Muñoz MA (2014). "Trophic coherence determines food-web stability". Proc Natl Acad Sci USA. 111 (50): 17923–17928. arXiv:1404.7728. Bibcode:2014PNAS..11117923J. doi:10.1073/pnas.1409077111. PMC 4273378. PMID 25468963.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Levine S (1980). "Several measures of trophic structure applicable to complex food webs". J Theor Biol. 83 (2): 195–207. doi:10.1016/0022-5193(80)90288-X.
- 1 2 3 4 5 6 Johnson S and Jones NS (2017). "Looplessness in networks is linked to trophic coherence". Proc Natl Acad Sci USA. 114 (22): 5618–5623. arXiv:1505.07332. doi:10.1073/pnas.1613786114. PMC 5465891. PMID 28512222.
- ↑ Klaise J and Johnson S (2017). "The origin of motif families in food webs". Scientific Reports. 7 (1): 16197. arXiv:1609.04318. Bibcode:2017NatSR...716197K. doi:10.1038/s41598-017-15496-1. PMC 5700930. PMID 29170384.
- ↑ Domı́nguez-Garcı́a V, Johnson S, Muñoz MA (2016). "Intervality and coherence in complex networks". Chaos. 26 (6): 065308. arXiv:1603.03767. Bibcode:2016Chaos..26f5308D. doi:10.1063/1.4953163. PMID 27368797. S2CID 16081869.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - 1 2 3 4 Klaise J and Johnson S (2016). "From neurons to epidemics: How trophic coherence affects spreading processes". Chaos. 26 (6): 065310. arXiv:1603.00670. Bibcode:2016Chaos..26f5310K. doi:10.1063/1.4953160. PMID 27368799. S2CID 205214650.
- ↑ Newman, MEJ (2003). "The structure and function of complex networks". SIAM Review. 45 (2): 167–256. arXiv:cond-mat/0303516. Bibcode:2003SIAMR..45..167N. doi:10.1137/S003614450342480. S2CID 221278130.
- ↑ Rossberg AG, Brännström A, Dieckmann U (2010). "Food-web structure in low- and high-dimensional trophic niche spaces". J R Soc Interface. 7 (53): 1735–1743. doi:10.1098/rsif.2010.0111. PMC 2988264. PMID 20462875.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Cohen JE and Newman CM (1985). "A stochastic theory of community food webs I. Models and aggregated data". Proc. R. Soc. B. 224 (1237): 421–448. Bibcode:1985RSPSB.224..421C. doi:10.1098/rspb.1985.0042. S2CID 52993453.
- 1 2 Stouffer DB, Camacho J, Amaral LAN (2006). "A robust measure of food web intervality". Proc Natl Acad Sci USA. 103 (50): 19015–19020. Bibcode:2006PNAS..10319015S. doi:10.1073/pnas.0603844103. PMC 1748169. PMID 17146055.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Williams RJ and Martinez ND (2000). "Simple rules yield complex food webs". Nature. 404 (6774): 180–183. Bibcode:2000Natur.404..180W. doi:10.1038/35004572. PMID 10724169. S2CID 205004984.