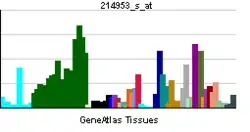
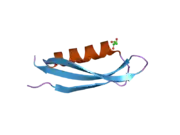
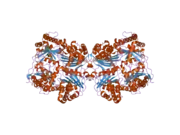Amyloid-beta precursor protein (APP) is an integral membrane protein expressed in many tissues and concentrated in the synapses of neurons. It functions as a cell surface receptor[5] and has been implicated as a regulator of synapse formation,[6] neural plasticity,[7] antimicrobial activity,[8] and iron export.[9] It is coded for by the gene APP and regulated by substrate presentation.[10] APP is best known as the precursor molecule whose proteolysis generates amyloid beta (Aβ), a polypeptide containing 37 to 49 amino acid residues, whose amyloid fibrillar form is the primary component of amyloid plaques found in the brains of Alzheimer's disease patients.
Genetics
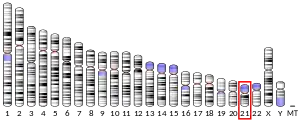
Amyloid-beta precursor protein is an ancient and highly conserved protein.[11] In humans, the gene APP is located on chromosome 21 and contains 18 exons spanning 290 kilobases.[12][13] Several alternative splicing isoforms of APP have been observed in humans, ranging in length from 639 to 770 amino acids, with certain isoforms preferentially expressed in neurons; changes in the neuronal ratio of these isoforms have been associated with Alzheimer's disease.[14] Homologous proteins have been identified in other organisms such as Drosophila (fruit flies), C. elegans (roundworms),[15] and all mammals.[16] The amyloid beta region of the protein, located in the membrane-spanning domain, is not well conserved across species and has no obvious connection with APP's native-state biological functions.[16]
Mutations in critical regions of amyloid precursor protein, including the region that generates amyloid beta (Aβ), cause familial susceptibility to Alzheimer's disease.[17][18][19][20] For example, several mutations outside the Aβ region associated with familial Alzheimer's have been found to dramatically increase production of Aβ.[21]
A mutation (A673T) in the APP gene protects against Alzheimer's disease. This substitution is adjacent to the beta secretase cleavage site and results in a 40% reduction in the formation of amyloid beta in vitro.[22]
Structure


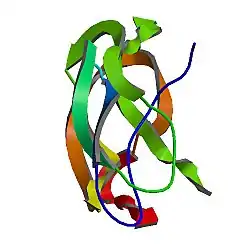
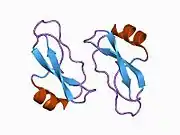
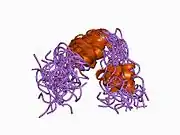
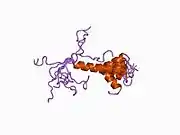
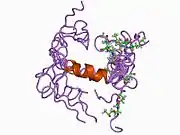
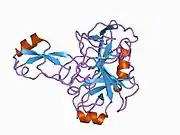
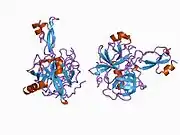
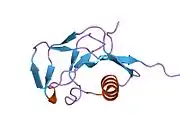
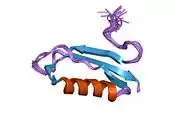
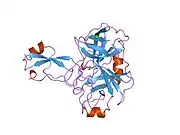
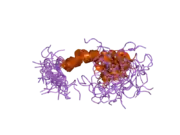
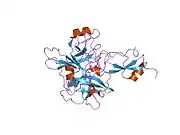
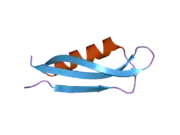
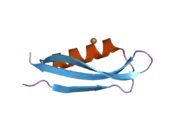
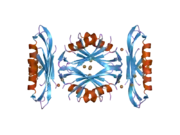
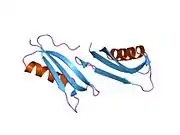
A number of different structural domains that fold mostly on their own have been found in the APP sequence. The extracellular region, much larger than the intracellular region, is divided into the E1 and E2 domains, linked by an acidic domain (AcD); E1 contains two subdomains including a growth factor-like domain (GFLD) and a copper-binding domain (CuBD) interacting tightly together.[24] A serine protease inhibitor domain, absent from the isoform differentially expressed in the brain, is found between acidic region and E2 domain.[25] The complete crystal structure of APP has not yet been solved; however, individual domains have been successfully crystallized, the growth factor-like domain,[26] the copper-binding domain,[27] the complete E1 domain[24] and the E2 domain.[23]
Post-translational processing
APP undergoes extensive post-translational modification including glycosylation, phosphorylation, sialylation, and tyrosine sulfation, as well as many types of proteolytic processing to generate peptide fragments.[28] It is commonly cleaved by proteases in the secretase family; alpha secretase and beta secretase both remove nearly the entire extracellular domain to release membrane-anchored carboxy-terminal fragments that may be associated with apoptosis.[16] Cleavage by gamma secretase within the membrane-spanning domain after beta-secretase cleavage generates the amyloid-beta fragment; gamma secretase is a large multi-subunit complex whose components have not yet been fully characterized, but include presenilin, whose gene has been identified as a major genetic risk factor for Alzheimer's.[29]
The amyloidogenic processing of APP has been linked to its presence in lipid rafts. When APP molecules occupy a lipid raft region of membrane, they are more accessible to and differentially cleaved by beta secretase, whereas APP molecules outside a raft are differentially cleaved by the non-amyloidogenic alpha secretase.[30] Gamma secretase activity has also been associated with lipid rafts.[31] The role of cholesterol in lipid raft maintenance has been cited as a likely explanation for observations that high cholesterol and apolipoprotein E genotype are major risk factors for Alzheimer's disease.[32]
Biological function
Although the native biological role of APP is of obvious interest to Alzheimer's research, thorough understanding has remained elusive. Experimental models of Alzheimer's disease are commonly used by researchers to gain better understandings about the biological function of APP in disease pathology and progression.
Synaptic formation and repair
The most-substantiated role for APP is in synaptic formation and repair;[6] its expression is upregulated during neuronal differentiation and after neural injury. Roles in cell signaling, long-term potentiation, and cell adhesion have been proposed and supported by as-yet limited research.[16] In particular, similarities in post-translational processing have invited comparisons to the signaling role of the surface receptor protein Notch.[33]
APP knockout mice are viable and have relatively minor phenotypic effects including impaired long-term potentiation and memory loss without general neuron loss.[34] On the other hand, transgenic mice with upregulated APP expression have also been reported to show impaired long-term potentiation.[35]
The logical inference is that because Aβ accumulates excessively in Alzheimer's disease its precursor, APP, would be elevated as well. However, neuronal cell bodies contain less APP as a function of their proximity to amyloid plaques.[36] The data indicate that this deficit in APP results from a decline in production rather than an increase in catalysis. Loss of a neuron's APP may affect physiological deficits that contribute to dementia.
Somatic recombination
In neurons of the human brain, somatic recombination occurs frequently in the gene that encodes APP.[37] Neurons from individuals with sporadic Alzheimer's disease show greater APP gene diversity due to somatic recombination than neurons from healthy individuals.[37]
Anterograde neuronal transport
Molecules synthesized in the cell bodies of neurons must be conveyed outward to the distal synapses. This is accomplished via fast anterograde transport. It has been found that APP can mediate interaction between cargo and kinesin and thus facilitate this transport. Specifically, a short peptide 15-amino-acid sequence from the cytoplasmic carboxy-terminus is necessary for interaction with the motor protein.[38]
Additionally, it has been shown that the interaction between APP and kinesin is specific to the peptide sequence of APP.[39] In a recent experiment involving transport of peptide-conjugated colored beads, controls were conjugated to a single amino acid, glycine, such that they display the same terminal carboxylic acid group as APP without the intervening 15-amino-acid sequence mentioned above. The control beads were not motile, which demonstrated that the terminal COOH moiety of peptides is not sufficient to mediate transport.
Iron export
A different perspective on Alzheimer's is revealed by a mouse study that has found that APP possesses ferroxidase activity similar to ceruloplasmin, facilitating iron export through interaction with ferroportin; it seems that this activity is blocked by zinc trapped by accumulated Aβ in Alzheimer's.[9] It has been shown that a single nucleotide polymorphism in the 5'UTR of APP mRNA can disrupt its translation.[40]
The hypothesis that APP has ferroxidase activity in its E2 domain and facilitates export of Fe(II) is possibly incorrect since the proposed ferroxidase site of APP located in the E2 domain does not have ferroxidase activity.[41][42]
As APP does not possess ferroxidase activity within its E2 domain, the mechanism of APP-modulated iron efflux from ferroportin has come under scrutiny. One model suggests that APP acts to stabilize the iron efflux protein ferroportin in the plasma membrane of cells thereby increasing the total number of ferroportin molecules at the membrane. These iron-transporters can then be activated by known mammalian ferroxidases (i.e. ceruloplasmin or hephaestin).[43]
Hormonal regulation
The amyloid-β precursor protein (AβPP), and all associated secretases, are expressed early in development and play a key role in the endocrinology of reproduction – with the differential processing of AβPP by secretases regulating human embryonic stem cell (hESC) proliferation as well as their differentiation into neural precursor cells (NPC). The pregnancy hormone human chorionic gonadotropin (hCG) increases AβPP expression[44] and hESC proliferation while progesterone directs AβPP processing towards the non-amyloidogenic pathway, which promotes hESC differentiation into NPC.[45][46][47]
AβPP and its cleavage products do not promote the proliferation and differentiation of post-mitotic neurons; rather, the overexpression of either wild-type or mutant AβPP in post-mitotic neurons induces apoptotic death following their re-entry into the cell cycle.[48] It is postulated that the loss of sex steroids (including progesterone) but the elevation in luteinizing hormone, the adult equivalent of hCG, post-menopause and during andropause drives amyloid-β production[49] and re-entry of post-mitotic neurons into the cell cycle.
Interactions
Amyloid precursor protein has been shown to interact with:
APP interacts with reelin, a protein implicated in a number of brain disorders, including Alzheimer's disease.[70]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000142192 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000022892 - Ensembl, May 2017
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Amyloid-beta precursor protein". Retrieved 10 January 2021.
- 1 2 Priller C, Bauer T, Mitteregger G, Krebs B, Kretzschmar HA, Herms J (Jul 2006). "Synapse formation and function is modulated by the amyloid precursor protein". The Journal of Neuroscience. 26 (27): 7212–21. doi:10.1523/JNEUROSCI.1450-06.2006. PMC 6673945. PMID 16822978.
- ↑ Turner PR, O'Connor K, Tate WP, Abraham WC (May 2003). "Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory". Progress in Neurobiology. 70 (1): 1–32. doi:10.1016/S0301-0082(03)00089-3. PMID 12927332. S2CID 25376584.
- ↑ Moir RD, Lathe R, Tanzi RE (2018). "The antimicrobial protection hypothesis of Alzheimer's disease". Alzheimer's & Dementia. 14 (12): 1602–1614. doi:10.1016/j.jalz.2018.06.3040. PMID 30314800.
- 1 2 Duce JA, Tsatsanis A, Cater MA, James SA, Robb E, Wikhe K, Leong SL, Perez K, Johanssen T, Greenough MA, Cho HH, Galatis D, Moir RD, Masters CL, McLean C, Tanzi RE, Cappai R, Barnham KJ, Ciccotosto GD, Rogers JT, Bush AI (Sep 2010). "Iron-export ferroxidase activity of β-amyloid precursor protein is inhibited by zinc in Alzheimer's disease". Cell. 142 (6): 857–67. doi:10.1016/j.cell.2010.08.014. PMC 2943017. PMID 20817278.
- ↑ Wang, Hao; Kulas, Joshua A.; Wang, Chao; Holtzman, David M.; Ferris, Heather A.; Hansen, Scott B. (17 August 2021). "Regulation of beta-amyloid production in neurons by astrocyte-derived cholesterol". Proceedings of the National Academy of Sciences. 118 (33): e2102191118. Bibcode:2021PNAS..11802191W. doi:10.1073/pnas.2102191118. PMC 8379952. PMID 34385305.
- ↑ Tharp WG, Sarkar IN (April 2013). "Origins of amyloid-β". BMC Genomics. 14 (1): 290. doi:10.1186/1471-2164-14-290. PMC 3660159. PMID 23627794.
- ↑ Yoshikai S, Sasaki H, Doh-ura K, Furuya H, Sakaki Y (Mar 1990). "Genomic organization of the human amyloid beta-protein precursor gene". Gene. 87 (2): 257–63. doi:10.1016/0378-1119(90)90310-N. PMID 2110105.
- ↑ Lamb BT, Sisodia SS, Lawler AM, Slunt HH, Kitt CA, Kearns WG, Pearson PL, Price DL, Gearhart JD (Sep 1993). "Introduction and expression of the 400 kilobase amyloid precursor protein gene in transgenic mice [corrected]". Nature Genetics. 5 (1): 22–30. doi:10.1038/ng0993-22. PMID 8220418. S2CID 42752531.
- ↑ Matsui T, Ingelsson M, Fukumoto H, Ramasamy K, Kowa H, Frosch MP, Irizarry MC, Hyman BT (Aug 2007). "Expression of APP pathway mRNAs and proteins in Alzheimer's disease". Brain Research. 1161: 116–23. doi:10.1016/j.brainres.2007.05.050. PMID 17586478. S2CID 26901380.
- ↑ Ewald, Collin Y.; Li, Chris (2012-04-01). "Caenorhabditis elegans as a model organism to study APP function". Experimental Brain Research. 217 (3–4): 397–411. doi:10.1007/s00221-011-2905-7. ISSN 0014-4819. PMC 3746071. PMID 22038715.
- 1 2 3 4 Zheng H, Koo EH (2006). "The amyloid precursor protein: beyond amyloid". Molecular Neurodegeneration. 1 (1): 5. doi:10.1186/1750-1326-1-5. PMC 1538601. PMID 16930452.
- ↑ Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L (Feb 1991). "Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease". Nature. 349 (6311): 704–6. Bibcode:1991Natur.349..704G. doi:10.1038/349704a0. PMID 1671712. S2CID 4336069.
- ↑ Murrell J, Farlow M, Ghetti B, Benson MD (Oct 1991). "A mutation in the amyloid precursor protein associated with hereditary Alzheimer's disease". Science. 254 (5028): 97–9. Bibcode:1991Sci...254...97M. doi:10.1126/science.1925564. PMID 1925564.
- ↑ Chartier-Harlin MC, Crawford F, Houlden H, Warren A, Hughes D, Fidani L, Goate A, Rossor M, Roques P, Hardy J (Oct 1991). "Early-onset Alzheimer's disease caused by mutations at codon 717 of the beta-amyloid precursor protein gene". Nature. 353 (6347): 844–6. Bibcode:1991Natur.353..844C. doi:10.1038/353844a0. PMID 1944558. S2CID 4345311.
- ↑ Lloyd, GM; Trejo-Lopez, JA; Xia, Y; McFarland, KN; Lincoln, SJ; Ertekin-Taner, N; Giasson, BI; Yachnis, AT; Prokop, S (12 March 2020). "Prominent amyloid plaque pathology and cerebral amyloid angiopathy in APP V717I (London) carrier - phenotypic variability in autosomal dominant Alzheimer's disease". Acta Neuropathologica Communications. 8 (1): 31. doi:10.1186/s40478-020-0891-3. PMC 7068954. PMID 32164763.
- ↑ Citron M, Oltersdorf T, Haass C, McConlogue L, Hung AY, Seubert P, Vigo-Pelfrey C, Lieberburg I, Selkoe DJ (Dec 1992). "Mutation of the beta-amyloid precursor protein in familial Alzheimer's disease increases beta-protein production". Nature. 360 (6405): 672–4. Bibcode:1992Natur.360..672C. doi:10.1038/360672a0. PMID 1465129. S2CID 4341170.
- ↑ Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, Hoyte K, Gustafson A, Liu Y, Lu Y, Bhangale T, Graham RR, Huttenlocher J, Bjornsdottir G, Andreassen OA, Jönsson EG, Palotie A, Behrens TW, Magnusson OT, Kong A, Thorsteinsdottir U, Watts RJ, Stefansson K (Aug 2012). "A mutation in APP protects against Alzheimer's disease and age-related cognitive decline". Nature. 488 (7409): 96–9. Bibcode:2012Natur.488...96J. doi:10.1038/nature11283. PMID 22801501. S2CID 4333449.
- 1 2 PDB: 1RW6; Wang Y, Ha Y (Aug 2004). "The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain". Molecular Cell. 15 (3): 343–53. doi:10.1016/j.molcel.2004.06.037. PMID 15304215.
- 1 2 Dahms SO, Hoefgen S, Roeser D, Schlott B, Gührs KH, Than ME (Mar 2010). "Structure and biochemical analysis of the heparin-induced E1 dimer of the amyloid precursor protein". Proceedings of the National Academy of Sciences of the United States of America. 107 (12): 5381–6. Bibcode:2010PNAS..107.5381D. doi:10.1073/pnas.0911326107. PMC 2851805. PMID 20212142.; see also PDB ID 3KTM
- ↑ Sisodia SS, Koo EH, Hoffman PN, Perry G, Price DL (Jul 1993). "Identification and transport of full-length amyloid precursor proteins in rat peripheral nervous system". The Journal of Neuroscience. 13 (7): 3136–42. doi:10.1523/JNEUROSCI.13-07-03136.1993. PMC 6576678. PMID 8331390.
- ↑ Rossjohn J, Cappai R, Feil SC, Henry A, McKinstry WJ, Galatis D, Hesse L, Multhaup G, Beyreuther K, Masters CL, Parker MW (Apr 1999). "Crystal structure of the N-terminal, growth factor-like domain of Alzheimer amyloid precursor protein". Nature Structural Biology. 6 (4): 327–31. doi:10.1038/7562. PMID 10201399. S2CID 30925432.; see also PDB ID 1MWP
- ↑ Kong GK, Adams JJ, Harris HH, Boas JF, Curtain CC, Galatis D, Masters CL, Barnham KJ, McKinstry WJ, Cappai R, Parker MW (Mar 2007). "Structural studies of the Alzheimer's amyloid precursor protein copper-binding domain reveal how it binds copper ions". Journal of Molecular Biology. 367 (1): 148–61. doi:10.1016/j.jmb.2006.12.041. PMID 17239395.; See also 2007 PDB IDs 2FJZ, 2FK2, 2FKL.
- ↑ De Strooper B, Annaert W (Jun 2000). "Proteolytic processing and cell biological functions of the amyloid precursor protein". Journal of Cell Science. 113 (11): 1857–70. doi:10.1242/jcs.113.11.1857. PMID 10806097.
- ↑ Chen F, Hasegawa H, Schmitt-Ulms G, Kawarai T, Bohm C, Katayama T, Gu Y, Sanjo N, Glista M, Rogaeva E, Wakutani Y, Pardossi-Piquard R, Ruan X, Tandon A, Checler F, Marambaud P, Hansen K, Westaway D, St George-Hyslop P, Fraser P (Apr 2006). "TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity". Nature. 440 (7088): 1208–12. doi:10.1038/nature04667. PMID 16641999. S2CID 4349251.
- ↑ Ehehalt R, Keller P, Haass C, Thiele C, Simons K (Jan 2003). "Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts". The Journal of Cell Biology. 160 (1): 113–23. doi:10.1083/jcb.200207113. PMC 2172747. PMID 12515826.
- ↑ Vetrivel KS, Cheng H, Lin W, Sakurai T, Li T, Nukina N, Wong PC, Xu H, Thinakaran G (Oct 2004). "Association of gamma-secretase with lipid rafts in post-Golgi and endosome membranes". The Journal of Biological Chemistry. 279 (43): 44945–54. doi:10.1074/jbc.M407986200. PMC 1201506. PMID 15322084.
- ↑ Riddell DR, Christie G, Hussain I, Dingwall C (Aug 2001). "Compartmentalization of beta-secretase (Asp2) into low-buoyant density, noncaveolar lipid rafts". Current Biology. 11 (16): 1288–93. doi:10.1016/S0960-9822(01)00394-3. PMID 11525745. S2CID 15502857.
- ↑ Selkoe D, Kopan R (2003). "Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration". Annual Review of Neuroscience. 26 (1): 565–97. doi:10.1146/annurev.neuro.26.041002.131334. PMID 12730322.
- ↑ Phinney AL, Calhoun ME, Wolfer DP, Lipp HP, Zheng H, Jucker M (1999). "No hippocampal neuron or synaptic bouton loss in learning-impaired aged beta-amyloid precursor protein-null mice". Neuroscience. 90 (4): 1207–16. doi:10.1016/S0306-4522(98)00645-9. PMID 10338291. S2CID 6001957.
- ↑ Matsuyama S, Teraoka R, Mori H, Tomiyama T (2007). "Inverse correlation between amyloid precursor protein and synaptic plasticity in transgenic mice". NeuroReport. 18 (10): 1083–7. doi:10.1097/WNR.0b013e3281e72b18. PMID 17558301. S2CID 34157306.
- ↑ Barger SW, DeWall KM, Liu L, Mrak RE, Griffin WS (Aug 2008). "Relationships between expression of apolipoprotein E and beta-amyloid precursor protein are altered in proximity to Alzheimer beta-amyloid plaques: potential explanations from cell culture studies". Journal of Neuropathology and Experimental Neurology. 67 (8): 773–83. doi:10.1097/NEN.0b013e318180ec47. PMC 3334532. PMID 18648325.
- 1 2 Lee MH, Siddoway B, Kaeser GE, Segota I, Rivera R, Romanow WJ, Liu CS, Park C, Kennedy G, Long T, Chun J (November 2018). "Somatic APP gene recombination in Alzheimer's disease and normal neurons". Nature. 563 (7733): 639–645. Bibcode:2018Natur.563..639L. doi:10.1038/s41586-018-0718-6. PMC 6391999. PMID 30464338.
- ↑ Satpute-Krishnan P, DeGiorgis JA, Conley MP, Jang M, Bearer EL (Oct 2006). "A peptide zipcode sufficient for anterograde transport within amyloid precursor protein". Proceedings of the National Academy of Sciences of the United States of America. 103 (44): 16532–7. Bibcode:2006PNAS..10316532S. doi:10.1073/pnas.0607527103. PMC 1621108. PMID 17062754.
- ↑ Seamster PE, Loewenberg M, Pascal J, Chauviere A, Gonzales A, Cristini V, Bearer EL (Oct 2012). "Quantitative measurements and modeling of cargo-motor interactions during fast transport in the living axon". Physical Biology. 9 (5): 055005. Bibcode:2012PhBio...9e5005S. doi:10.1088/1478-3975/9/5/055005. PMC 3625656. PMID 23011729.
- ↑ Rogers JT, Bush AI, Cho HH, Smith DH, Thomson AM, Friedlich AL, Lahiri DK, Leedman PJ, Huang X, Cahill CM (Dec 2008). "Iron and the translation of the amyloid precursor protein (APP) and ferritin mRNAs: riboregulation against neural oxidative damage in Alzheimer's disease". Biochemical Society Transactions. 36 (Pt 6): 1282–7. doi:10.1042/BST0361282. PMC 2746665. PMID 19021541.
- ↑ Ebrahimi KH, Hagedoorn PL, Hagen WR (2012). "A synthetic peptide with the putative iron binding motif of amyloid precursor protein (APP) does not catalytically oxidize iron". PLOS ONE. 7 (8): e40287. Bibcode:2012PLoSO...740287E. doi:10.1371/journal.pone.0040287. PMC 3419245. PMID 22916096.
- ↑ Honarmand Ebrahimi K, Dienemann C, Hoefgen S, Than ME, Hagedoorn PL, Hagen WR (2013). "The amyloid precursor protein (APP) does not have a ferroxidase site in its E2 domain". PLOS ONE. 8 (8): e72177. Bibcode:2013PLoSO...872177H. doi:10.1371/journal.pone.0072177. PMC 3747053. PMID 23977245.
- ↑ McCarthy RC, Park YH, Kosman DJ (Jul 2014). "sAPP modulates iron efflux from brain microvascular endothelial cells by stabilizing the ferrous iron exporter ferroportin". EMBO Reports. 15 (7): 809–15. doi:10.15252/embr.201338064. PMC 4196985. PMID 24867889.
- ↑ Porayette P, Gallego MJ, Kaltcheva MM, Meethal SV, Atwood CS (Dec 2007). "Amyloid-beta precursor protein expression and modulation in human embryonic stem cells: a novel role for human chorionic gonadotropin". Biochemical and Biophysical Research Communications. 364 (3): 522–7. doi:10.1016/j.bbrc.2007.10.021. PMID 17959150.
- ↑ Porayette P, Gallego MJ, Kaltcheva MM, Bowen RL, Vadakkadath Meethal S, Atwood CS (Aug 2009). "Differential processing of amyloid-beta precursor protein directs human embryonic stem cell proliferation and differentiation into neuronal precursor cells". The Journal of Biological Chemistry. 284 (35): 23806–17. doi:10.1074/jbc.M109.026328. PMC 2749153. PMID 19542221.
- ↑ Gallego MJ, Porayette P, Kaltcheva MM, Meethal SV, Atwood CS (Jun 2009). "Opioid and progesterone signaling is obligatory for early human embryogenesis". Stem Cells and Development. 18 (5): 737–40. doi:10.1089/scd.2008.0190. PMC 2891507. PMID 18803462.
- ↑ Gallego MJ, Porayette P, Kaltcheva MM, Bowen RL, Vadakkadath Meethal S, Atwood CS (2010). "The pregnancy hormones human chorionic gonadotropin and progesterone induce human embryonic stem cell proliferation and differentiation into neuroectodermal rosettes". Stem Cell Research & Therapy. 1 (4): 28. doi:10.1186/scrt28. PMC 2983441. PMID 20836886.
- ↑ McPhie DL, Coopersmith R, Hines-Peralta A, Chen Y, Ivins KJ, Manly SP, Kozlowski MR, Neve KA, Neve RL (Jul 2003). "DNA synthesis and neuronal apoptosis caused by familial Alzheimer disease mutants of the amyloid precursor protein are mediated by the p21 activated kinase PAK3". The Journal of Neuroscience. 23 (17): 6914–27. doi:10.1523/JNEUROSCI.23-17-06914.2003. PMC 6740729. PMID 12890786.
- ↑ Bowen RL, Verdile G, Liu T, Parlow AF, Perry G, Smith MA, Martins RN, Atwood CS (May 2004). "Luteinizing hormone, a reproductive regulator that modulates the processing of amyloid-beta precursor protein and amyloid-beta deposition". The Journal of Biological Chemistry. 279 (19): 20539–45. doi:10.1074/jbc.M311993200. PMID 14871891.
- 1 2 3 Biederer T, Cao X, Südhof TC, Liu X (Sep 2002). "Regulation of APP-dependent transcription complexes by Mint/X11s: differential functions of Mint isoforms". The Journal of Neuroscience. 22 (17): 7340–51. doi:10.1523/JNEUROSCI.22-17-07340.2002. PMC 6757996. PMID 12196555.
- 1 2 Borg JP, Ooi J, Levy E, Margolis B (Nov 1996). "The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein". Molecular and Cellular Biology. 16 (11): 6229–41. doi:10.1128/mcb.16.11.6229. PMC 231626. PMID 8887653.
- 1 2 Araki Y, Tomita S, Yamaguchi H, Miyagi N, Sumioka A, Kirino Y, Suzuki T (Dec 2003). "Novel cadherin-related membrane proteins, Alcadeins, enhance the X11-like protein-mediated stabilization of amyloid beta-protein precursor metabolism". The Journal of Biological Chemistry. 278 (49): 49448–58. doi:10.1074/jbc.M306024200. PMID 12972431.
- ↑ Tomita S, Ozaki T, Taru H, Oguchi S, Takeda S, Yagi Y, Sakiyama S, Kirino Y, Suzuki T (Jan 1999). "Interaction of a neuron-specific protein containing PDZ domains with Alzheimer's amyloid precursor protein". The Journal of Biological Chemistry. 274 (4): 2243–54. doi:10.1074/jbc.274.4.2243. PMID 9890987.
- ↑ Tanahashi H, Tabira T (Feb 1999). "X11L2, a new member of the X11 protein family, interacts with Alzheimer's beta-amyloid precursor protein". Biochemical and Biophysical Research Communications. 255 (3): 663–7. doi:10.1006/bbrc.1999.0265. PMID 10049767.
- ↑ Zambrano N, Buxbaum JD, Minopoli G, Fiore F, De Candia P, De Renzis S, Faraonio R, Sabo S, Cheetham J, Sudol M, Russo T (Mar 1997). "Interaction of the phosphotyrosine interaction/phosphotyrosine binding-related domains of Fe65 with wild-type and mutant Alzheimer's beta-amyloid precursor proteins". The Journal of Biological Chemistry. 272 (10): 6399–405. doi:10.1074/jbc.272.10.6399. PMID 9045663.
- ↑ Guénette SY, Chen J, Jondro PD, Tanzi RE (Oct 1996). "Association of a novel human FE65-like protein with the cytoplasmic domain of the beta-amyloid precursor protein". Proceedings of the National Academy of Sciences of the United States of America. 93 (20): 10832–7. Bibcode:1996PNAS...9310832G. doi:10.1073/pnas.93.20.10832. PMC 38241. PMID 8855266.
- ↑ Tanahashi H, Tabira T (Feb 1999). "Molecular cloning of human Fe65L2 and its interaction with the Alzheimer's beta-amyloid precursor protein". Neuroscience Letters. 261 (3): 143–6. doi:10.1016/S0304-3940(98)00995-1. PMID 10081969. S2CID 54307954.
- ↑ Trommsdorff M, Borg JP, Margolis B, Herz J (Dec 1998). "Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein". The Journal of Biological Chemistry. 273 (50): 33556–60. doi:10.1074/jbc.273.50.33556. PMID 9837937.
- ↑ Chow N, Korenberg JR, Chen XN, Neve RL (May 1996). "APP-BP1, a novel protein that binds to the carboxyl-terminal region of the amyloid precursor protein". The Journal of Biological Chemistry. 271 (19): 11339–46. doi:10.1074/jbc.271.19.11339. PMID 8626687.
- ↑ Zheng P, Eastman J, Vande Pol S, Pimplikar SW (Dec 1998). "PAT1, a microtubule-interacting protein, recognizes the basolateral sorting signal of amyloid precursor protein". Proceedings of the National Academy of Sciences of the United States of America. 95 (25): 14745–50. Bibcode:1998PNAS...9514745Z. doi:10.1073/pnas.95.25.14745. PMC 24520. PMID 9843960.
- ↑ Wang B, Nguyen M, Breckenridge DG, Stojanovic M, Clemons PA, Kuppig S, Shore GC (Apr 2003). "Uncleaved BAP31 in association with A4 protein at the endoplasmic reticulum is an inhibitor of Fas-initiated release of cytochrome c from mitochondria". The Journal of Biological Chemistry. 278 (16): 14461–8. doi:10.1074/jbc.M209684200. PMID 12529377.
- ↑ Lefterov IM, Koldamova RP, Lazo JS (Sep 2000). "Human bleomycin hydrolase regulates the secretion of amyloid precursor protein". FASEB Journal. 14 (12): 1837–47. doi:10.1096/fj.99-0938com. PMID 10973933. S2CID 44302063.
- ↑ Araki Y, Miyagi N, Kato N, Yoshida T, Wada S, Nishimura M, Komano H, Yamamoto T, De Strooper B, Yamamoto K, Suzuki T (Jun 2004). "Coordinated metabolism of Alcadein and amyloid beta-protein precursor regulates FE65-dependent gene transactivation". The Journal of Biological Chemistry. 279 (23): 24343–54. doi:10.1074/jbc.M401925200. PMID 15037614.
- ↑ Ikezu T, Trapp BD, Song KS, Schlegel A, Lisanti MP, Okamoto T (Apr 1998). "Caveolae, plasma membrane microdomains for alpha-secretase-mediated processing of the amyloid precursor protein". The Journal of Biological Chemistry. 273 (17): 10485–95. doi:10.1074/jbc.273.17.10485. PMID 9553108.
- ↑ Hashimoto T, Wakabayashi T, Watanabe A, Kowa H, Hosoda R, Nakamura A, Kanazawa I, Arai T, Takio K, Mann DM, Iwatsubo T (Apr 2002). "CLAC: a novel Alzheimer amyloid plaque component derived from a transmembrane precursor, CLAC-P/collagen type XXV". The EMBO Journal. 21 (7): 1524–34. doi:10.1093/emboj/21.7.1524. PMC 125364. PMID 11927537.
- ↑ Ohsawa I, Takamura C, Kohsaka S (Mar 2001). "Fibulin-1 binds the amino-terminal head of beta-amyloid precursor protein and modulates its physiological function". Journal of Neurochemistry. 76 (5): 1411–20. doi:10.1046/j.1471-4159.2001.00144.x. PMID 11238726. S2CID 83321033.
- ↑ Chauhan VP, Ray I, Chauhan A, Wisniewski HM (May 1999). "Binding of gelsolin, a secretory protein, to amyloid beta-protein". Biochemical and Biophysical Research Communications. 258 (2): 241–6. doi:10.1006/bbrc.1999.0623. PMID 10329371.
- ↑ Yan SD, Fu J, Soto C, Chen X, Zhu H, Al-Mohanna F, Collison K, Zhu A, Stern E, Saido T, Tohyama M, Ogawa S, Roher A, Stern D (Oct 1997). "An intracellular protein that binds amyloid-beta peptide and mediates neurotoxicity in Alzheimer's disease". Nature. 389 (6652): 689–95. Bibcode:1997Natur.389..689D. doi:10.1038/39522. PMID 9338779. S2CID 4343238.
- ↑ Tarr PE, Roncarati R, Pelicci G, Pelicci PG, D'Adamio L (May 2002). "Tyrosine phosphorylation of the beta-amyloid precursor protein cytoplasmic tail promotes interaction with Shc". The Journal of Biological Chemistry. 277 (19): 16798–804. doi:10.1074/jbc.M110286200. PMID 11877420.
- ↑ Hoe HS, Lee KJ, Carney RS, Lee J, Markova A, Lee JY, Howell BW, Hyman BT, Pak DT, Bu G, Rebeck GW (Jun 2009). "Interaction of reelin with amyloid precursor protein promotes neurite outgrowth". The Journal of Neuroscience. 29 (23): 7459–73. doi:10.1523/JNEUROSCI.4872-08.2009. PMC 2759694. PMID 19515914.
Further reading
- Beyreuther K, Pollwein P, Multhaup G, Mönning U, König G, Dyrks T, Schubert W, Masters CL (Sep 1993). "Regulation and expression of the Alzheimer's beta/A4 amyloid protein precursor in health, disease, and Down's syndrome". Annals of the New York Academy of Sciences. 695 (1 Transduction): 91–102. doi:10.1111/j.1749-6632.1993.tb23035.x. PMID 8239320. S2CID 22058428.
- Straub JE, Guevara J, Huo S, Lee JP (Jun 2002). "Long time dynamic simulations: exploring the folding pathways of an Alzheimer's amyloid Abeta-peptide". Accounts of Chemical Research. 35 (6): 473–81. doi:10.1021/ar010031e. PMID 12069633.
- Annaert W, De Strooper B (2003). "A cell biological perspective on Alzheimer's disease". Annual Review of Cell and Developmental Biology. 18 (1): 25–51. doi:10.1146/annurev.cellbio.18.020402.142302. PMID 12142279.
- Koo EH (Nov 2002). "The beta-amyloid precursor protein (APP) and Alzheimer's disease: does the tail wag the dog?". Traffic. 3 (11): 763–70. doi:10.1034/j.1600-0854.2002.31101.x. PMID 12383342. S2CID 40411806.
- Van Nostrand WE, Melchor JP, Romanov G, Zeigler K, Davis J (Nov 2002). "Pathogenic effects of cerebral amyloid angiopathy mutations in the amyloid beta-protein precursor". Annals of the New York Academy of Sciences. 977 (1): 258–65. Bibcode:2002NYASA.977..258N. doi:10.1111/j.1749-6632.2002.tb04824.x. PMID 12480759. S2CID 22567664.
- Ling Y, Morgan K, Kalsheker N (Nov 2003). "Amyloid precursor protein (APP) and the biology of proteolytic processing: relevance to Alzheimer's disease". The International Journal of Biochemistry & Cell Biology. 35 (11): 1505–35. doi:10.1016/S1357-2725(03)00133-X. PMID 12824062.
- Kerr ML, Small DH (Apr 2005). "Cytoplasmic domain of the beta-amyloid protein precursor of Alzheimer's disease: function, regulation of proteolysis, and implications for drug development". Journal of Neuroscience Research. 80 (2): 151–9. doi:10.1002/jnr.20408. PMID 15672415. S2CID 31985212.
- Maynard CJ, Bush AI, Masters CL, Cappai R, Li QX (Jun 2005). "Metals and amyloid-beta in Alzheimer's disease". International Journal of Experimental Pathology. 86 (3): 147–59. doi:10.1111/j.0959-9673.2005.00434.x. PMC 2517409. PMID 15910549.
- Tickler AK, Wade JD, Separovic F (Aug 2005). "The role of Abeta peptides in Alzheimer's disease". Protein and Peptide Letters. 12 (6): 513–9. doi:10.2174/0929866054395905. PMID 16101387.
- Reinhard C, Hébert SS, De Strooper B (Dec 2005). "The amyloid-beta precursor protein: integrating structure with biological function". The EMBO Journal. 24 (23): 3996–4006. doi:10.1038/sj.emboj.7600860. PMC 1356301. PMID 16252002.
- Watson D, Castaño E, Kokjohn TA, Kuo YM, Lyubchenko Y, Pinsky D, Connolly ES, Esh C, Luehrs DC, Stine WB, Rowse LM, Emmerling MR, Roher AE (Dec 2005). "Physicochemical characteristics of soluble oligomeric Abeta and their pathologic role in Alzheimer's disease". Neurological Research. 27 (8): 869–81. doi:10.1179/016164105X49436. hdl:11336/43663. PMID 16354549. S2CID 25687818.
- Calinisan V, Gravem D, Chen RP, Brittin S, Mohandas N, Lecomte MC, Gascard P (2006). "New insights into potential functions for the protein 4.1 superfamily of proteins in kidney epithelium". Frontiers in Bioscience. 11 (1): 1646–66. doi:10.2741/1911. PMID 16368544. S2CID 26325962.
- Vetrivel KS, Thinakaran G (Jan 2006). "Amyloidogenic processing of beta-amyloid precursor protein in intracellular compartments". Neurology. 66 (2 Suppl 1): S69–73. doi:10.1212/01.wnl.0000192107.17175.39. PMID 16432149. S2CID 35965729.
- Gallo C, Orlassino R, Vineis C (Feb 2006). "[Recurrent intraparenchimal haemorrhages in a patient with cerebral amyloidotic angiopathy: description of one autopsy case]". Pathologica. 98 (1): 44–7. PMID 16789686.
- Coulson EJ (Aug 2006). "Does the p75 neurotrophin receptor mediate Abeta-induced toxicity in Alzheimer's disease?". Journal of Neurochemistry. 98 (3): 654–60. doi:10.1111/j.1471-4159.2006.03905.x. PMID 16893414. S2CID 20879380.
- Menéndez-González M, Pérez-Pinera P, Martínez-Rivera M, Calatayud MT, Blázquez Menes B (2006). "APP processing and the APP-KPI domain involvement in the amyloid cascade". Neuro-Degenerative Diseases. 2 (6): 277–83. doi:10.1159/000092315. PMID 16909010. S2CID 45002038.
- Neve RL, McPhie DL (Apr 2007). "Dysfunction of amyloid precursor protein signaling in neurons leads to DNA synthesis and apoptosis". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1772 (4): 430–7. doi:10.1016/j.bbadis.2006.10.008. PMC 1862818. PMID 17113271.
- Chen X, Stern D, Yan SD (Dec 2006). "Mitochondrial dysfunction and Alzheimer's disease". Current Alzheimer Research. 3 (5): 515–20. doi:10.2174/156720506779025215. PMID 17168650.
- Caltagarone J, Jing Z, Bowser R (Apr 2007). "Focal adhesions regulate Abeta signaling and cell death in Alzheimer's disease". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1772 (4): 438–45. doi:10.1016/j.bbadis.2006.11.007. PMC 1876750. PMID 17215111.
- Wolfe MS (Feb 2007). "When loss is gain: reduced presenilin proteolytic function leads to increased Abeta42/Abeta40. Talking Point on the role of presenilin mutations in Alzheimer disease". EMBO Reports. 8 (2): 136–40. doi:10.1038/sj.embor.7400896. PMC 1796780. PMID 17268504.
External links
- GeneReviews/NCBI/NIH/UW entry on Early-Onset Familial Alzheimer Disease
- Amyloid+Protein+Precursor at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Entrez Gene: APP amyloid beta (A4) precursor protein (peptidase nexin-II, Alzheimer disease)
- Human APP genome location and APP gene details page in the UCSC Genome Browser.