 | ||
major histocompatibility complex (human), class I, B7 | ||
| Alleles | B*0702, *0703, *0704, *0705 | |
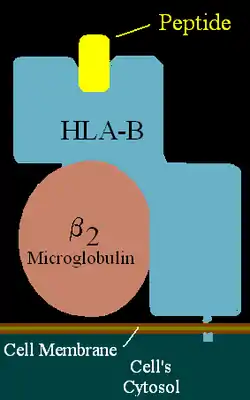
| Structure (See HLA-B) | ||
| Alleles (See Serotyping) | ||
| Locus | chr.6 6p21.31 | |
HLA-B7 (B7) is an HLA-B serotype. The serotype identifies the more common HLA-B*07 gene products.[1] (For terminology help see: HLA-serotype tutorial) B7, previously HL-A7, was one of the first 'HL-A' antigens recognized, largely because of the frequency of B*0702 in Northern and Western Europe and the United States. B7 is found in two major haplotypes in Europe, where it reaches peak frequency in Ireland. One haplotype A3-B7-DR15-DQ1 can be found over a vast region and is in apparent selective disequilibrium. B7 is a risk factor for cervical cancer, sarcoidosis, and early-onset spondylarthropathies.
Serology
| B*07 | B7 | Sample |
| allele | % | size (N) |
| *0702 | 98 | 10841 |
| *0703 | 93 | 15 |
| *0704 | 89 | 44 |
| *0705 | 95 | 42 |
| *0706 | 96 | 23 |
| *0707 | 92 | 13 |
| *0709 | 78 | 9 |
| Alleles link-out to IMGT/HLA Databease at EBI | ||
Alleles
| freq | ||
| ref. | Population | (%) |
| [3] | Ireland South | 17.6 |
| [3] | Ireland Northern | 17.3 |
| [3] | Australia New South Wales | 12.0 |
| [3] | Croatia | 9.7 |
| [3] | Azores S. Maria & Miguel | 9.0 |
| [3] | Cameroon Beti | 8.6 |
| [3] | Saudi Arabia Guraiat and Hail | 8.3 |
| [3] | Azores Central Islands | 8.0 |
| [3] | France South East | 7.2 |
| [3] | Cameroon Bamileke | 7.1 |
| [3] | Portugal Centre | 7.0 |
| [3] | Italy North pop 1 | 6.7 |
| [3] | Japan Central | 6.5 |
| [3] | Czech Republic | 6.1 |
| [3] | Uganda Kampala | 5.9 |
| [3] | Mali Bandiagara | 5.8 |
| [3] | Senegal Niokholo Mandenka | 5.8 |
| [3] | India Mumbai Marathas | 4.9 |
| [3] | Zambia Lusaka | 4.6 |
| [3] | Zimbabwe Harare Shona | 4.6 |
| [3] | South African Natal Zulu | 4.5 |
| [3] | Romanian | 3.7 |
| [3] | South Korea (3) | 3.5 |
| [3] | Shijiazhuang Tianjian Han, China | 3.4 |
| [3] | India North Delhi | 3.3 |
| [3] | Kenya Luo | 2.5 |
| [3] | China Guangzhou Han | 2.4 |
| [3] | Mexico Chihuahua Tarahumara | 2.3 |
| [3] | Sudanese | 2.3 |
| [3] | Singapore Javanese Indonesians | 2.0 |
| [3] | Spain Eastern Andalusia Gipsy | 2.0 |
| [3] | New Caledonia | 1.9 |
| [3] | Oman | 1.7 |
| [3] | USA Alaska Yupik Natives | 1.6 |
| [3] | China Beijing | 1.5 |
| [3] | Tunisia | 1.5 |
| [3] | Argentina Toba Rosario | 1.2 |
| [3] | Singapore Chinese Han | 1.2 |
| [3] | USA Arizona Pima | 1.1 |
| [3] | American Samoa | 1.0 |
| [3] | Japan Ainu Hokkaido | 1.0 |
| [3] | Kenya Nandi | 1.0 |
| [3] | Portugal South | 1.0 |
| [3] | Singapore Riau Malay | 1.0 |
| [3] | Singapore Thai | 1.0 |
In disease
Cervical cancer
HLA-B7 along with HLA-DQ8 increased risk for cervical cancer in Costa Rican [4] and South Asian women[5]
Sarcoidosis
A weak relationship between HLA-B7 and sarcoidosis has been known for 30+ years, [6] yet has not consistently been reproducible in all studies however. A common serologically defined haplotype in Europeans is HLA A3-Cw7-B7-DR15-DQ6.2 which is composed of alleles A*0301:Cw*0701:B*0702:DRB1*1501:DQA1*0102:DQB1*0602. In persistent sarcoidosis this haplotype appears elevated, further study eliminated risk contributed by A3-Cw7 and DQ6.2 indicating B7-DR15 haplotype contains risk of disease (OR = 2.5). Corresponding region of chromosome 6 contains nearly one million nucleotides thus these genes, or another closely linked gene could be involved in such massing of inflammatory granulomata.[7]
Juvenile Spondylarthropathies
In Croatian children, two HLA-B27 alleles were found associated with disease, B*2702, B*2705.[8] The study showed also B*0702 in cooperation with B*27, the HLA-B*07/B*27 combination with D6S273-134 genomic marker allele and was found not to be the result of linkage disequilibrium. B*2705 was found to be dominant allele associated.
Haemochromatosis
The HFE gene responsible for haemochromatosis is distal on chromosome 6 from HLA-A and more so from HLA-B, the distance suffices (3 million nucleotides) to allow equilibration of the loci. Nonetheless, a linkage has been found between A3-B7 haplotype and haemochromatosis. The region is almost 1.4 million nucleotides in length and contains many other genes that could be involved. A more recent study looked at a number of linked gene-alleles and found I82-2:D6S265-1:HLA-A3:D6S128-2:HLA-F1:D6S105-8 was constantly associated while B7 appeared beyond the haplotype linked to disease.[9]
Covid-19
In october 2021, a team of researcher from Centre hospitalier universitaire Sainte-Justine in Montreal, Canada, announced the discovery of HLA-B7 genetic marker as a potential cause for severe form of covid-19. While they noted that more work will be necessary to confirm this discovery, they found that individuals carrying the HLA-B7 genetic marker, which represents 35% of the population worldwide, are more likely to have a less effective immune response to covid-19.[10][11]
References
- ↑ Marsh, S. G.; Albert, E. D.; Bodmer, W. F.; Bontrop, R. E.; Dupont, B.; Erlich, H. A.; Fernández-Viña, M.; Geraghty, D. E.; Holdsworth, R.; Hurley, C. K.; Lau, M.; Lee, K. W.; Mach, B.; Maiers, M.; Mayr, W. R.; Müller, C. R.; Parham, P.; Petersdorf, E. W.; Sasazuki, T.; Strominger, J. L.; Svejgaard, A.; Terasaki, P. I.; Tiercy, J. M.; Trowsdale, J. (2010). "Nomenclature for factors of the HLA system, 2010". Tissue Antigens. 75 (4): 291–455. doi:10.1111/j.1399-0039.2010.01466.x. PMC 2848993. PMID 20356336.
- ↑ derived from IMGT/HLA
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 Middleton D, Menchaca L, Rood H, Komerofsky R (2003). "New allele frequency database: http://www.allelefrequencies.net". Tissue Antigens. 61 (5): 403–7. doi:10.1034/j.1399-0039.2003.00062.x. PMID 12753660.
{{cite journal}}: External link in|title= - ↑ Wang SS, Wheeler CM, Hildesheim A, et al. (November 2001). "Human leukocyte antigen class I and II alleles and risk of cervical neoplasia: results from a population-based study in Costa Rica". J. Infect. Dis. 184 (10): 1310–4. doi:10.1086/324209. PMID 11679920.
- ↑ Bhattacharya P, Sengupta S (October 2007). "Predisposition to HPV16/18-related cervical cancer because of proline homozygosity at codon 72 of p53 among Indian women is influenced by HLA-B*07 and homozygosity of HLA-DQB1*03". Tissue Antigens. 70 (4): 283–93. doi:10.1111/j.1399-0039.2007.00894.x. PMID 17767549.
- ↑ Rybicki BA, Iannuzzi MC (March 2004). "Sarcoidosis and human leukocyte antigen class I and II genes: it takes two to tango?". Am. J. Respir. Crit. Care Med. 169 (6): 665–6. doi:10.1164/rccm.2401005. PMID 15003948.
- ↑ Grunewald J, Eklund A, Olerup O (March 2004). "Human leukocyte antigen class I alleles and the disease course in sarcoidosis patients". Am. J. Respir. Crit. Care Med. 169 (6): 696–702. CiteSeerX 10.1.1.321.2788. doi:10.1164/rccm.200303-459OC. PMID 14656748.
- ↑ Harjacek M, Margetić T, Kerhin-Brkljacić V, Martinez N, Grubić Z (2008). "HLA-B*27/HLA-B*07 in combination with D6S273-134 allele is associated with increased susceptibility to juvenile spondyloarthropathies". Clin. Exp. Rheumatol. 26 (3): 498–504. PMID 18578977.
- ↑ Raha-Chowdhury R, Bowen DJ, Burnett AK, Worwood M (June 1995). "Allelic associations and homozygosity at loci from HLA-B to D6S299 in genetic haemochromatosis". J. Med. Genet. 32 (6): 446–52. doi:10.1136/jmg.32.6.446. PMC 1050484. PMID 7666396.
- ↑ "Une découverte québécoise pourrait permettre d'identifier les patients à risque d'une forme grave de la COVID-19". 12 October 2021.
- ↑ https://ici.radio-canada.ca/nouvelle/1830925/covid-identification-rapide-patients-risque-maladie-grave/