Heat shock 70 kDa protein 1, also termed Hsp72, is a protein that in humans is encoded by the HSPA1A gene.[5][6] As a member of the heat shock protein 70 family and a chaperone protein, it facilitates the proper folding of newly translated and misfolded proteins, as well as stabilize or degrade mutant proteins.[5][6] In addition, Hsp72 also facilitates DNA repair.[7] Its functions contribute to biological processes including signal transduction, apoptosis, protein homeostasis, and cell growth and differentiation.[6][8] It has been associated with an extensive number of cancers, neurodegenerative diseases, cell senescence and aging, and inflammatory diseases such as Diabetes mellitus type 2 and rheumatoid arthritis.[9][10][8]
Structure
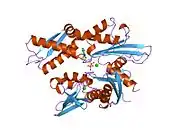
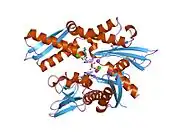
This intronless gene encodes a 70kDa heat shock protein which is a member of the heat shock protein 70 (Hsp70) family.[5] As a Hsp70 protein, it has a C-terminal protein substrate-binding domain and an N-terminal ATP-binding domain.[11][12][13] The substrate-binding domain consists of two subdomains, a two-layered β-sandwich subdomain (SBDβ) and an α-helical subdomain (SBDα), which are connected by the loop Lα,β. SBDβ contains the peptide binding pocket while SBDα serves as a lid to cover the substrate binding cleft. The ATP binding domain consists of four subdomains split into two lobes by a central ATP/ADP binding pocket. The two terminal domains are linked together by a conserved region referred to as loop LL,1, which is critical for allosteric regulation. The unstructured region at the very end of the C-terminal is believed to be the docking site for co-chaperones.[13]
Function
This protein is a member of the Hsp70 family. In conjunction with other heat shock proteins, this protein stabilizes existing proteins against aggregation and mediates the folding of newly translated proteins in the cytosol and in organelles.[5] In order to properly fold non-native proteins, this protein interacts with the hydrophobic peptide segments of proteins in an ATP-controlled fashion. Though the exact mechanism still remains unclear, there are at least two alternative modes of action: kinetic partitioning and local unfolding. In kinetic partitioning, Hsp70s repetitively bind and release substrates in cycles that maintain low concentrations of free substrate. This effectively prevents aggregation while allowing free molecules to fold to the native state. In local unfolding, the binding and release cycles induce localized unfolding in the substrate, which helps to overcome kinetic barriers for folding to the native state.[6] Ultimately, its role in protein folding contributes to its function in signal transduction, apoptosis, protein homeostasis, and cell growth and differentiation.[6][8]
In addition to the process of protein folding, transport and degradation, this Hsp70 member can preserve the function of mutant proteins. Nonetheless, effects of these mutations can still manifest when Hsp70 chaperones are overwhelmed during stress conditions.[6] Hsp72 also protects against DNA damage and participates in DNA repair, including base excision repair (BER) and nucleotide excision repair (NER).[7] Furthermore, this protein enhances antigen-specific tumor immunity by facilitating more efficient antigen presentation to cytotoxic T cells.[8] It is also involved in the ubiquitin-proteasome pathway through interaction with the AU-rich element RNA-binding protein 1. The gene is located in the major histocompatibility complex class III region, in a cluster with two closely related genes which encode similar proteins.[5] Finally, Hsp72 can protect against disrupted metabolic homeostasis by inducing production of pro-inflammatory cytokines, tumor necrosis factor-α, interleukin 1β, and interleukin-6 in immune cells, thereby reducing inflammation and improving skeletal muscle oxidation.[9][14] Though at very low levels under normal conditions, HSP72 expression greatly increases under stress, effectively protecting cells from adverse effects in various pathological states.[15]
Along with its role in DNA repair, Hsp72 is also directly involved in caspase-dependent apoptosis by binding Apaf-1, thereby inhibiting procaspase-9 activation and release of cytochrome c.[11] Additionally, Hsp72 has been observed to inhibit apoptosis by preventing the release of SMAC/Diablo and binding XIAP to prevent its degradation.[12] Hsp72 is also involved in caspase-independent apoptosis, as it also binds AIFM1.[11]
Clinical significance
The Hsp70 member proteins are important apoptotic constituents. During a normal embryologic processes, or during cell injury (such as ischemia-reperfusion injury during heart attacks and strokes) or during developments and processes in cancer, an apoptotic cell undergoes structural changes including cell shrinkage, plasma membrane blebbing, nuclear condensation, and fragmentation of the DNA and nucleus. This is followed by fragmentation into apoptotic bodies that are quickly removed by phagocytes, thereby preventing an inflammatory response.[16] It is a mode of cell death defined by characteristic morphological, biochemical and molecular changes. It was first described as a "shrinkage necrosis", and then this term was replaced by apoptosis to emphasize its role opposite mitosis in tissue kinetics. In later stages of apoptosis the entire cell becomes fragmented, forming a number of plasma membrane-bounded apoptotic bodies which contain nuclear and or cytoplasmic elements. The ultrastructural appearance of necrosis is quite different, the main features being mitochondrial swelling, plasma membrane breakdown and cellular disintegration. Apoptosis occurs in many physiological and pathological processes. It plays an important role during embryonal development as programmed cell death and accompanies a variety of normal involutional processes in which it serves as a mechanism to remove "unwanted" cells.
Hsp70 member proteins, including Hsp72, inhibit apoptosis by acting on the caspase-dependent pathway and against apoptosis-inducing agents such as tumor necrosis factor-α (TNFα), staurosporine, and doxorubicin. This role leads to its involvement in many pathological processes, such as oncogenesis, neurodegeneration, and senescence. In particular, overexpression of HSP72 has been linked to the development some cancers, such as hepatocellular carcinoma, gastric cancers, colon cancers, breast cancers, and lung cancers, which led to its use as a prognostic marker for these cancers.[8] Elevated Hsp70 levels in tumor cells may increase malignancy and resistance to therapy by complexing, and hence, stabilizing, oncofetal proteins and products and transporting them into intracellular sites, thereby promoting tumor cell proliferation.[6][8] As a result, tumor vaccine strategies for Hsp70s have been highly successful in animal models and progressed to clinical trials.[8] One treatment, a Hsp72/AFP recombined vaccine, elicited robust protective immunity against AFP-expressing tumors in mice experiments. Therefore, the vaccine holds promise for treating hepatocellular carcinoma.[8] Alternatively, overexpression of Hsp70 can mitigate damage from ischemia-reperfusion in cardiac muscle, as well damage from neurodegenerative diseases, such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, and spinocerebellar ataxias, and aging and cell senescence, as observed in centenarians subjected to heat shock challenge.[6][17]
In Diabetes mellitus type 2 (T2DM), a small molecule activator of Hsp72 named BGP-15 has been shown to improve insulin sensitivity and inflammation in an insulin-resistant mouse model, increase mitochondrial volume, and improve metabolic homeostasis in a rat model of T2DM. BGP-15 has now proceeded to Phase 2b clinical trials and demonstrated no side-effects thus far. Though early speculation considered that Hsp72 expression might be affecting insulin sensitivity through a direct interaction with GLUT4, studies were unable to verify this link. Experiments did reveal that Hsp72 improved insulin sensitivity through stimulating glucose uptake during a hyperinsulemic-euglycemic clamp in T2DM patients.[9] Additionally, Hsp72 has been associated with another inflammatory condition, rheumatoid arthritis, and could be implemented to help diagnose and monitor disease activity in patients.[10]
Interactions
HSPA1A has been shown to interact with:
See also
References
- 1 2 3 ENSG00000204389, ENSG00000237724, ENSG00000235941, ENSG00000215328 GRCh38: Ensembl release 89: ENSG00000234475, ENSG00000204389, ENSG00000237724, ENSG00000235941, ENSG00000215328 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000091971 - Ensembl, May 2017
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- 1 2 3 4 5 "Entrez Gene: HSPA1A heat shock 70kDa protein 1A".
- 1 2 3 4 5 6 7 8 9 Mayer MP, Bukau B (Mar 2005). "Hsp70 chaperones: cellular functions and molecular mechanism". Cellular and Molecular Life Sciences. 62 (6): 670–684. doi:10.1007/s00018-004-4464-6. PMC 2773841. PMID 15770419.
- 1 2 3 4 Duan Y, Huang S, Yang J, Niu P, Gong Z, Liu X, Xin L, Currie RW, Wu T (Mar 2014). "HspA1A facilitates DNA repair in human bronchial epithelial cells exposed to Benzo[a]pyrene and interacts with casein kinase 2". Cell Stress & Chaperones. 19 (2): 271–9. doi:10.1007/s12192-013-0454-7. PMC 3933616. PMID 23979991.
- 1 2 3 4 5 6 7 8 Wang X, Wang Q, Lin H, Li S, Sun L, Yang Y (Feb 2013). "HSP72 and gp96 in gastroenterological cancers". Clinica Chimica Acta; International Journal of Clinical Chemistry. 417: 73–9. doi:10.1016/j.cca.2012.12.017. PMID 23266770.
- 1 2 3 Henstridge DC, Whitham M, Febbraio MA (Nov 2014). "Chaperoning to the metabolic party: The emerging therapeutic role of heat-shock proteins in obesity and type 2 diabetes". Molecular Metabolism. 3 (8): 781–93. doi:10.1016/j.molmet.2014.08.003. PMC 4216407. PMID 25379403.
- 1 2 Najafizadeh SR, Ghazizadeh Z, Nargesi AA, Mahdavi M, Abtahi S, Mirmiranpour H, Nakhjavani M (Mar 2015). "Analysis of serum heat shock protein 70 (HSPA1A) concentrations for diagnosis and disease activity monitoring in patients with rheumatoid arthritis". Cell Stress & Chaperones. 20 (3): 537–43. doi:10.1007/s12192-015-0578-z. PMC 4406931. PMID 25739548.
- 1 2 3 4 5 Ravagnan L, Gurbuxani S, Susin SA, Maisse C, Daugas E, Zamzami N, Mak T, Jäättelä M, Penninger JM, Garrido C, Kroemer G (September 2001). "Heat-shock protein 70 antagonizes apoptosis-inducing factor". Nat. Cell Biol. 3 (9): 839–43. doi:10.1038/ncb0901-839. PMID 11533664. S2CID 21164493.
- 1 2 3 Zhang B, Rong R, Li H, Peng X, Xiong L, Wang Y, Yu X, Mao H (2015). "Heat shock protein 72 suppresses apoptosis by increasing the stability of X-linked inhibitor of apoptosis protein in renal ischemia/reperfusion injury". Mol Med Rep. 11 (3): 1793–9. doi:10.3892/mmr.2014.2939. PMC 4270332. PMID 25394481.
- 1 2 Zhang P, Leu JI, Murphy ME, George DL, Marmorstein R (2014). "Crystal structure of the stress-inducible human heat shock protein 70 substrate-binding domain in complex with peptide substrate". PLOS ONE. 9 (7): e103518. Bibcode:2014PLoSO...9j3518Z. doi:10.1371/journal.pone.0103518. PMC 4110032. PMID 25058147.
- ↑ Gibson OR, Dennis A, Parfitt T, Taylor L, Watt PW, Maxwell NS (May 2014). "Extracellular Hsp72 concentration relates to a minimum endogenous criteria during acute exercise-heat exposure". Cell Stress & Chaperones. 19 (3): 389–400. doi:10.1007/s12192-013-0468-1. PMC 3982022. PMID 24085588.
- ↑ Ryu DS, Yang H, Lee SE, Park CS, Jin YH, Park YS (Nov 2013). "Crotonaldehyde induces heat shock protein 72 expression that mediates anti-apoptotic effects in human endothelial cells". Toxicology Letters. 223 (2): 116–23. doi:10.1016/j.toxlet.2013.09.010. PMID 24070736.
- ↑ Kerr JF, Wyllie AH, Currie AR (Aug 1972). "Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics". British Journal of Cancer. 26 (4): 239–57. doi:10.1038/bjc.1972.33. PMC 2008650. PMID 4561027.
- ↑ Henstridge DC, Whitham M, Febbraio MA (2014). "Chaperoning to the metabolic party: The emerging therapeutic role of heat-shock proteins in obesity and type 2 diabetes". Mol Metab. 3 (8): 781–93. doi:10.1016/j.molmet.2014.08.003. PMC 4216407. PMID 25379403.
- ↑ Ruchalski K, Mao H, Singh SK, Wang Y, Mosser DD, Li F, Schwartz JH, Borkan SC (December 2003). "HSP72 inhibits apoptosis-inducing factor release in ATP-depleted renal epithelial cells". Am. J. Physiol., Cell Physiol. 285 (6): C1483–93. doi:10.1152/ajpcell.00049.2003. PMID 12930708.
- ↑ Park HS, Cho SG, Kim CK, Hwang HS, Noh KT, Kim MS, Huh SH, Kim MJ, Ryoo K, Kim EK, Kang WJ, Lee JS, Seo JS, Ko YG, Kim S, Choi EJ (November 2002). "Heat shock protein hsp72 is a negative regulator of apoptosis signal-regulating kinase 1". Mol. Cell. Biol. 22 (22): 7721–30. doi:10.1128/mcb.22.22.7721-7730.2002. PMC 134722. PMID 12391142.
- ↑ Doong H, Price J, Kim YS, Gasbarre C, Probst J, Liotta LA, Blanchette J, Rizzo K, Kohn E (September 2000). "CAIR-1/BAG-3 forms an EGF-regulated ternary complex with phospholipase C-gamma and Hsp70/Hsc70". Oncogene. 19 (38): 4385–95. doi:10.1038/sj.onc.1203797. PMID 10980614. S2CID 28684205.
- ↑ Antoku K, Maser RS, Scully WJ, Delach SM, Johnson DE (September 2001). "Isolation of Bcl-2 binding proteins that exhibit homology with BAG-1 and suppressor of death domains protein". Biochem. Biophys. Res. Commun. 286 (5): 1003–10. doi:10.1006/bbrc.2001.5512. PMID 11527400.
- ↑ Pang Q, Christianson TA, Keeble W, Koretsky T, Bagby GC (December 2002). "The anti-apoptotic function of Hsp70 in the interferon-inducible double-stranded RNA-dependent protein kinase-mediated death signaling pathway requires the Fanconi anemia protein, FANCC". J. Biol. Chem. 277 (51): 49638–43. doi:10.1074/jbc.M209386200. PMID 12397061.
- ↑ Reuter TY, Medhurst AL, Waisfisz Q, Zhi Y, Herterich S, Hoehn H, Gross HJ, Joenje H, Hoatlin ME, Mathew CG, Huber PA (October 2003). "Yeast two-hybrid screens imply involvement of Fanconi anemia proteins in transcription regulation, cell signaling, oxidative metabolism, and cellular transport". Exp. Cell Res. 289 (2): 211–21. doi:10.1016/s0014-4827(03)00261-1. PMID 14499622.
- 1 2 3 Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, Takahashi R (July 2002). "CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity". Mol. Cell. 10 (1): 55–67. doi:10.1016/s1097-2765(02)00583-x. PMID 12150907.
- ↑ Shi Y, Mosser DD, Morimoto RI (March 1998). "Molecular chaperones as HSF1-specific transcriptional repressors". Genes Dev. 12 (5): 654–66. doi:10.1101/gad.12.5.654. PMC 316571. PMID 9499401.
- ↑ Zhou X, Tron VA, Li G, Trotter MJ (August 1998). "Heat shock transcription factor-1 regulates heat shock protein-72 expression in human keratinocytes exposed to ultraviolet B light". J. Invest. Dermatol. 111 (2): 194–8. doi:10.1046/j.1523-1747.1998.00266.x. PMID 9699716.
- ↑ Nakamura T, Hinagata J, Tanaka T, Imanishi T, Wada Y, Kodama T, Doi T (January 2002). "HSP90, HSP70, and GAPDH directly interact with the cytoplasmic domain of macrophage scavenger receptors". Biochem. Biophys. Res. Commun. 290 (2): 858–64. doi:10.1006/bbrc.2001.6271. PMID 11785981.
- ↑ Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C (June 1999). "Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions". Mol. Cell. Biol. 19 (6): 4535–45. doi:10.1128/mcb.19.6.4535. PMC 104411. PMID 10330192.
Further reading
- Andersen JL, Planelles V (2005). "The role of Vpr in HIV-1 pathogenesis". Curr. HIV Res. 3 (1): 43–51. doi:10.2174/1570162052772988. PMID 15638722.
- Zhao RY, Elder RT (2005). "Viral infections and cell cycle G2/M regulation". Cell Res. 15 (3): 143–9. doi:10.1038/sj.cr.7290279. PMID 15780175.
- Zhao RY, Bukrinsky M, Elder RT (2005). "HIV-1 viral protein R (Vpr) & host cellular responses". Indian J. Med. Res. 121 (4): 270–86. PMID 15817944.
- Muthumani K, Choo AY, Premkumar A, Hwang DS, Thieu KP, Desai BM, Weiner DB (2006). "Human immunodeficiency virus type 1 (HIV-1) Vpr-regulated cell death: insights into mechanism". Cell Death Differ. 12 (Suppl 1): 962–70. doi:10.1038/sj.cdd.4401583. PMID 15832179.
- Grosz MD, Womack JE, Skow LC (1993). "Syntenic conservation of HSP70 genes in cattle and humans". Genomics. 14 (4): 863–8. doi:10.1016/S0888-7543(05)80106-5. PMID 1478667.
- Veldscholte J, Berrevoets CA, Brinkmann AO, Grootegoed JA, Mulder E (1992). "Anti-androgens and the mutated androgen receptor of LNCaP cells: differential effects on binding affinity, heat-shock protein interaction, and transcription activation". Biochemistry. 31 (8): 2393–9. doi:10.1021/bi00123a026. PMID 1540595.
- Abravaya K, Myers MP, Murphy SP, Morimoto RI (1992). "The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression". Genes Dev. 6 (7): 1153–64. doi:10.1101/gad.6.7.1153. PMID 1628823.
- Milner CM, Campbell RD (1990). "Structure and expression of the three MHC-linked HSP70 genes". Immunogenetics. 32 (4): 242–51. doi:10.1007/BF00187095. PMID 1700760. S2CID 9531492.
- Sargent CA, Dunham I, Trowsdale J, Campbell RD (1989). "Human major histocompatibility complex contains genes for the major heat shock protein HSP70". Proc. Natl. Acad. Sci. U.S.A. 86 (6): 1968–72. Bibcode:1989PNAS...86.1968S. doi:10.1073/pnas.86.6.1968. PMC 286826. PMID 2538825.
- Wu B, Hunt C, Morimoto R (1985). "Structure and expression of the human gene encoding major heat shock protein HSP70". Mol. Cell. Biol. 5 (2): 330–41. doi:10.1128/MCB.5.2.330. PMC 366716. PMID 2858050.
- Goate AM, Cooper DN, Hall C, Leung TK, Solomon E, Lim L (1987). "Localization of a human heat-shock HSP 70 gene sequence to chromosome 6 and detection of two other loci by somatic-cell hybrid and restriction fragment length polymorphism analysis". Hum. Genet. 75 (2): 123–8. doi:10.1007/BF00591072. PMID 2880793. S2CID 33788452.
- Hickey E, Brandon SE, Sadis S, Smale G, Weber LA (1986). "Molecular cloning of sequences encoding the human heat-shock proteins and their expression during hyperthermia". Gene. 43 (1–2): 147–54. doi:10.1016/0378-1119(86)90018-1. PMID 3019832.
- Harrison GS, Drabkin HA, Kao FT, Hartz J, Hart IM, Chu EH, Wu BJ, Morimoto RI (1987). "Chromosomal location of human genes encoding major heat-shock protein HSP70" (PDF). Somat. Cell Mol. Genet. 13 (2): 119–30. doi:10.1007/BF01534692. hdl:2027.42/45535. PMID 3470951. S2CID 16069170.
- Drabent B, Genthe A, Benecke BJ (1987). "In vitro transcription of a human hsp 70 heat shock gene by extracts prepared from heat-shocked and non-heat-shocked human cells". Nucleic Acids Res. 14 (22): 8933–48. doi:10.1093/nar/14.22.8933. PMC 311921. PMID 3786141.
- Hunt C, Morimoto RI (1985). "Conserved features of eukaryotic hsp70 genes revealed by comparison with the nucleotide sequence of human hsp70". Proc. Natl. Acad. Sci. U.S.A. 82 (19): 6455–9. Bibcode:1985PNAS...82.6455H. doi:10.1073/pnas.82.19.6455. PMC 390735. PMID 3931075.
- Liao J, Lowthert LA, Ghori N, Omary MB (1995). "The 70-kDa heat shock proteins associate with glandular intermediate filaments in an ATP-dependent manner". J. Biol. Chem. 270 (2): 915–22. doi:10.1074/jbc.270.2.915. PMID 7529764.
- Selkirk JK, Merrick BA, Stackhouse BL, He C (1995). "Multiple p53 protein isoforms and formation of oligomeric complexes with heat shock proteins Hsp70 and Hsp90 in the human mammary tumor, T47D, cell line". Appl. Theor. Electrophor. 4 (1): 11–8. PMID 7811761.
- Furlini G, Vignoli M, Re MC, Gibellini D, Ramazzotti E, Zauli G, La Placa M (1994). "Human immunodeficiency virus type 1 interaction with the membrane of CD4+ cells induces the synthesis and nuclear translocation of 70K heat shock protein". J. Gen. Virol. 75 (1): 193–9. doi:10.1099/0022-1317-75-1-193. PMID 7906708.
- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Prapapanich V, Chen S, Toran EJ, Rimerman RA, Smith DF (1996). "Mutational analysis of the hsp70-interacting protein Hip". Mol. Cell. Biol. 16 (11): 6200–7. doi:10.1128/MCB.16.11.6200. PMC 231623. PMID 8887650.
External links
- HSPA1A+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Overview of all the structural information available in the PDB for UniProt: P0DMV8 (Heat shock 70 kDa protein 1A) at the PDBe-KB.
- Overview of all the structural information available in the PDB for UniProt: P0DMV9 (Heat shock 70 kDa protein 1B) at the PDBe-KB.