| Yeast | |
|---|---|
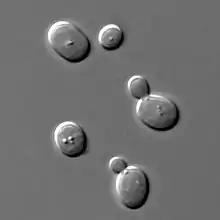 | |
| Saccharomyces cerevisiae, a species of yeast | |
.svg.png.webp) | |
| Cross-sectional labelled diagram of a typical yeast cell | |
| Scientific classification | |
| Domain: | Eukaryota |
| Kingdom: | Fungi |
| Phyla and subphyla with yeast species | |
Basidiomycota p. p. | |
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized.[1][2][3] They are estimated to constitute 1% of all described fungal species.[4]
Some yeast species have the ability to develop multicellular characteristics by forming strings of connected budding cells known as pseudohyphae or false hyphae, or quickly evolve into a multicellular cluster with specialised cell organelles function.[5][6] Yeast sizes vary greatly, depending on species and environment, typically measuring 3–4 µm in diameter, although some yeasts can grow to 40 µm in size.[7] Most yeasts reproduce asexually by mitosis, and many do so by the asymmetric division process known as budding. With their single-celled growth habit, yeasts can be contrasted with molds, which grow hyphae. Fungal species that can take both forms (depending on temperature or other conditions) are called dimorphic fungi.
The yeast species Saccharomyces cerevisiae converts carbohydrates to carbon dioxide and alcohols through the process of fermentation. The products of this reaction have been used in baking and the production of alcoholic beverages for thousands of years.[8] S. cerevisiae is also an important model organism in modern cell biology research, and is one of the most thoroughly studied eukaryotic microorganisms. Researchers have cultured it in order to understand the biology of the eukaryotic cell and ultimately human biology in great detail.[9] Other species of yeasts, such as Candida albicans, are opportunistic pathogens and can cause infections in humans. Yeasts have recently been used to generate electricity in microbial fuel cells[10] and to produce ethanol for the biofuel industry.
Yeasts do not form a single taxonomic or phylogenetic grouping. The term "yeast" is often taken as a synonym for Saccharomyces cerevisiae,[11] but the phylogenetic diversity of yeasts is shown by their placement in two separate phyla: the Ascomycota and the Basidiomycota. The budding yeasts or "true yeasts" are classified in the order Saccharomycetales,[12] within the phylum Ascomycota.
History
The word "yeast" comes from Old English gist, gyst, and from the Indo-European root yes-, meaning "boil", "foam", or "bubble".[13] Yeast microbes are probably one of the earliest domesticated organisms. Archaeologists digging in Egyptian ruins found early grinding stones and baking chambers for yeast-raised bread, as well as drawings of 4,000-year-old bakeries and breweries.[14] Vessels studied from several archaeological sites in Israel (dating to around 5,000, 3,000 and 2,500 years ago), which were believed to have contained alcoholic beverages (beer and mead), were found to contain yeast colonies that had survived over the millennia, providing the first direct biological evidence of yeast use in early cultures.[15] In 1680, Dutch naturalist Anton van Leeuwenhoek first microscopically observed yeast, but at the time did not consider them to be living organisms, but rather globular structures[16] as researchers were doubtful whether yeasts were algae or fungi.[17] Theodor Schwann recognized them as fungi in 1837.[18][19]
In 1857, French microbiologist Louis Pasteur showed that by bubbling oxygen into the yeast broth, cell growth could be increased, but fermentation was inhibited – an observation later called the "Pasteur effect". In the paper "Mémoire sur la fermentation alcoolique," Pasteur proved that alcoholic fermentation was conducted by living yeasts and not by a chemical catalyst.[14][20]
By the late 18th century two yeast strains used in brewing had been identified: Saccharomyces cerevisiae (top-fermenting yeast) and S. pastorianus (bottom-fermenting yeast). S. cerevisiae has been sold commercially by the Dutch for bread-making since 1780; while, around 1800, the Germans started producing S. cerevisiae in the form of cream. In 1825, a method was developed to remove the liquid so the yeast could be prepared as solid blocks.[21] The industrial production of yeast blocks was enhanced by the introduction of the filter press in 1867. In 1872, Baron Max de Springer developed a manufacturing process to create granulated yeast, a technique that was used until the first World War.[22] In the United States, naturally occurring airborne yeasts were used almost exclusively until commercial yeast was marketed at the Centennial Exposition in 1876 in Philadelphia, where Charles L. Fleischmann exhibited the product and a process to use it, as well as serving the resultant baked bread.[23]
The mechanical refrigerator (first patented in the 1850s in Europe) liberated brewers and winemakers from seasonal constraints for the first time and allowed them to exit cellars and other earthen environments. For John Molson, who made his livelihood in Montreal prior to the development of the fridge, the brewing season lasted from September through to May. The same seasonal restrictions formerly governed the distiller's art.[24]
Nutrition and growth
Yeasts are chemoorganotrophs, as they use organic compounds as a source of energy and do not require sunlight to grow. Carbon is obtained mostly from hexose sugars, such as glucose and fructose, or disaccharides such as sucrose and maltose. Some species can metabolize pentose sugars such as ribose,[25] alcohols, and organic acids. Yeast species either require oxygen for aerobic cellular respiration (obligate aerobes) or are anaerobic, but also have aerobic methods of energy production (facultative anaerobes). Unlike bacteria, no known yeast species grow only anaerobically (obligate anaerobes). Most yeasts grow best in a neutral or slightly acidic pH environment.
Yeasts vary in regard to the temperature range in which they grow best. For example, Leucosporidium frigidum grows at −2 to 20 °C (28 to 68 °F), Saccharomyces telluris at 5 to 35 °C (41 to 95 °F), and Candida slooffi at 28 to 45 °C (82 to 113 °F).[26] The cells can survive freezing under certain conditions, with viability decreasing over time.
In general, yeasts are grown in the laboratory on solid growth media or in liquid broths. Common media used for the cultivation of yeasts include potato dextrose agar or potato dextrose broth, Wallerstein Laboratories nutrient agar, yeast peptone dextrose agar, and yeast mould agar or broth. Home brewers who cultivate yeast frequently use dried malt extract and agar as a solid growth medium. The fungicide cycloheximide is sometimes added to yeast growth media to inhibit the growth of Saccharomyces yeasts and select for wild/indigenous yeast species. This will change the yeast process.
The appearance of a white, thready yeast, commonly known as kahm yeast, is often a byproduct of the lactofermentation (or pickling) of certain vegetables. It is usually the result of exposure to air. Although harmless, it can give pickled vegetables a bad flavor and must be removed regularly during fermentation.[27]
Ecology
Yeasts are very common in the environment, and are often isolated from sugar-rich materials. Examples include naturally occurring yeasts on the skins of fruits and berries (such as grapes, apples, or peaches), and exudates from plants (such as plant saps or cacti). Some yeasts are found in association with soil and insects.[28][29] Yeasts from the soil and from the skins of fruits and berries have been shown to dominate fungal succession during fruit decay.[30] The ecological function and biodiversity of yeasts are relatively unknown compared to those of other microorganisms.[31] Yeasts, including Candida albicans, Rhodotorula rubra, Torulopsis and Trichosporon cutaneum, have been found living in between people's toes as part of their skin flora.[32] Yeasts are also present in the gut flora of mammals and some insects[33] and even deep-sea environments host an array of yeasts.[34][35]
An Indian study of seven bee species and nine plant species found 45 species from 16 genera colonize the nectaries of flowers and honey stomachs of bees. Most were members of the genus Candida; the most common species in honey stomachs was Dekkera intermedia and in flower nectaries, Candida blankii.[36] Yeast colonising nectaries of the stinking hellebore have been found to raise the temperature of the flower, which may aid in attracting pollinators by increasing the evaporation of volatile organic compounds.[31][37] A black yeast has been recorded as a partner in a complex relationship between ants, their mutualistic fungus, a fungal parasite of the fungus and a bacterium that kills the parasite. The yeast has a negative effect on the bacteria that normally produce antibiotics to kill the parasite, so may affect the ants' health by allowing the parasite to spread.[38]
Certain strains of some species of yeasts produce proteins called yeast killer toxins that allow them to eliminate competing strains. (See main article on killer yeast.) This can cause problems for winemaking but could potentially also be used to advantage by using killer toxin-producing strains to make the wine. Yeast killer toxins may also have medical applications in treating yeast infections (see "Pathogenic yeasts" section below).[39]
Marine yeasts, defined as the yeasts that are isolated from marine environments, are able to grow better on a medium prepared using seawater rather than freshwater.[40] The first marine yeasts were isolated by Bernhard Fischer in 1894 from the Atlantic Ocean, and those were identified as Torula sp. and Mycoderma sp.[41] Following this discovery, various other marine yeasts have been isolated from around the world from different sources, including seawater, seaweeds, marine fish and mammals.[42] Among these isolates, some marine yeasts originated from terrestrial habitats (grouped as facultative marine yeast), which were brought to and survived in marine environments. The other marine yeasts were grouped as obligate or indigenous marine yeasts, which are confined to marine habitats.[41] However, no sufficient evidence has been found to explain the indispensability of seawater for obligate marine yeasts.[40] It has been reported that marine yeasts are able to produce many bioactive substances, such as amino acids, glucans, glutathione, toxins, enzymes, phytase, and vitamins with potential applications in the food, pharmaceutical, cosmetic, and chemical industries as well as for marine culture and environmental protection.[40] Marine yeast was successfully used to produce bioethanol using seawater-based media which will potentially reduce the water footprint of bioethanol.[43]
Reproduction
- Budding
- Conjugation
- Spore
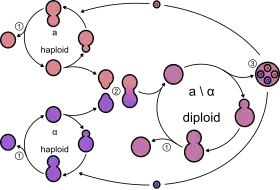
Yeasts, like all fungi, may have asexual and sexual reproductive cycles. The most common mode of vegetative growth in yeast is asexual reproduction by budding,[44] where a small bud (also known as a bleb or daughter cell) is formed on the parent cell. The nucleus of the parent cell splits into a daughter nucleus and migrates into the daughter cell. The bud then continues to grow until it separates from the parent cell, forming a new cell.[45] The daughter cell produced during the budding process is generally smaller than the mother cell. Some yeasts, including Schizosaccharomyces pombe, reproduce by fission instead of budding,[44] and thereby creating two identically sized daughter cells.
In general, under high-stress conditions such as nutrient starvation, haploid cells will die; under the same conditions, however, diploid cells can undergo sporulation, entering sexual reproduction (meiosis) and producing a variety of haploid spores, which can go on to mate (conjugate), reforming the diploid.[46]
The haploid fission yeast Schizosaccharomyces pombe is a facultative sexual microorganism that can undergo mating when nutrients are limited.[3][47] Exposure of S. pombe to hydrogen peroxide, an agent that causes oxidative stress leading to oxidative DNA damage, strongly induces mating and the formation of meiotic spores.[48] The budding yeast Saccharomyces cerevisiae reproduces by mitosis as diploid cells when nutrients are abundant, but when starved, this yeast undergoes meiosis to form haploid spores.[49] Haploid cells may then reproduce asexually by mitosis. Katz Ezov et al.[50] presented evidence that in natural S. cerevisiae populations clonal reproduction and selfing (in the form of intratetrad mating) predominate. In nature, the mating of haploid cells to form diploid cells is most often between members of the same clonal population and out-crossing is uncommon.[51] Analysis of the ancestry of natural S. cerevisiae strains led to the conclusion that out-crossing occurs only about once every 50,000 cell divisions.[51] These observations suggest that the possible long-term benefits of outcrossing (e.g. generation of diversity) are likely to be insufficient for generally maintaining sex from one generation to the next. Rather, a short-term benefit, such as recombinational repair during meiosis,[52] may be the key to the maintenance of sex in S. cerevisiae.
Some pucciniomycete yeasts, in particular species of Sporidiobolus and Sporobolomyces, produce aerially dispersed, asexual ballistoconidia.[53]
Uses
The useful physiological properties of yeast have led to their use in the field of biotechnology. Fermentation of sugars by yeast is the oldest and largest application of this technology. Many types of yeasts are used for making many foods: baker's yeast in bread production, brewer's yeast in beer fermentation, and yeast in wine fermentation and for xylitol production.[54] So-called red rice yeast is actually a mold, Monascus purpureus. Yeasts include some of the most widely used model organisms for genetics and cell biology.[55]
Alcoholic beverages
Alcoholic beverages are defined as beverages that contain ethanol (C2H5OH). This ethanol is almost always produced by fermentation – the metabolism of carbohydrates by certain species of yeasts under anaerobic or low-oxygen conditions. Beverages such as mead, wine, beer, or distilled spirits all use yeast at some stage of their production. A distilled beverage is a beverage containing ethanol that has been purified by distillation. Carbohydrate-containing plant material is fermented by yeast, producing a dilute solution of ethanol in the process. Spirits such as whiskey and rum are prepared by distilling these dilute solutions of ethanol. Components other than ethanol are collected in the condensate, including water, esters, and other alcohols, which (in addition to that provided by the oak in which it may be aged) account for the flavour of the beverage.
Beer


Brewing yeasts may be classed as "top-cropping" (or "top-fermenting") and "bottom-cropping" (or "bottom-fermenting").[56] Top-cropping yeasts are so called because they form a foam at the top of the wort during fermentation. An example of a top-cropping yeast is Saccharomyces cerevisiae, sometimes called an "ale yeast".[57] Bottom-cropping yeasts are typically used to produce lager-type beers, though they can also produce ale-type beers. These yeasts ferment well at low temperatures. An example of bottom-cropping yeast is Saccharomyces pastorianus, formerly known as S. carlsbergensis.
Decades ago, taxonomists reclassified S. carlsbergensis (uvarum) as a member of S. cerevisiae, noting that the only distinct difference between the two is metabolic. Lager strains of S. cerevisiae secrete an enzyme called melibiase, allowing them to hydrolyse melibiose, a disaccharide, into more fermentable monosaccharides. Top- and bottom-cropping and cold- and warm-fermenting distinctions are largely generalizations used by laypersons to communicate to the general public.[58]
The most common top-cropping brewer's yeast, S. cerevisiae, is the same species as the common baking yeast.[59] Brewer's yeast is also very rich in essential minerals and the B vitamins (except B12), a feature exploited in food products made from leftover (by-product) yeast from brewing.[60] However, baking and brewing yeasts typically belong to different strains, cultivated to favour different characteristics: baking yeast strains are more aggressive, to carbonate dough in the shortest amount of time possible; brewing yeast strains act more slowly but tend to produce fewer off-flavours and tolerate higher alcohol concentrations (with some strains, up to 22%).
Dekkera/Brettanomyces is a genus of yeast known for its important role in the production of 'lambic' and specialty sour ales, along with the secondary conditioning of a particular Belgian Trappist beer.[61] The taxonomy of the genus Brettanomyces has been debated since its early discovery and has seen many reclassifications over the years. Early classification was based on a few species that reproduced asexually (anamorph form) through multipolar budding.[62] Shortly after, the formation of ascospores was observed and the genus Dekkera, which reproduces sexually (teleomorph form), was introduced as part of the taxonomy.[63] The current taxonomy includes five species within the genera of Dekkera/Brettanomyces. Those are the anamorphs Brettanomyces bruxellensis, Brettanomyces anomalus, Brettanomyces custersianus, Brettanomyces naardenensis, and Brettanomyces nanus, with teleomorphs existing for the first two species, Dekkera bruxellensis and Dekkera anomala.[64] The distinction between Dekkera and Brettanomyces is arguable, with Oelofse et al. (2008) citing Loureiro and Malfeito-Ferreira from 2006 when they affirmed that current molecular DNA detection techniques have uncovered no variance between the anamorph and teleomorph states. Over the past decade, Brettanomyces spp. have seen an increasing use in the craft-brewing sector of the industry, with a handful of breweries having produced beers that were primarily fermented with pure cultures of Brettanomyces spp. This has occurred out of experimentation, as very little information exists regarding pure culture fermentative capabilities and the aromatic compounds produced by various strains. Dekkera/Brettanomyces spp. have been the subjects of numerous studies conducted over the past century, although a majority of the recent research has focused on enhancing the knowledge of the wine industry. Recent research on eight Brettanomyces strains available in the brewing industry focused on strain-specific fermentations and identified the major compounds produced during pure culture anaerobic fermentation in wort.[65]
Wine

Yeast is used in winemaking, where it converts the sugars present (glucose and fructose) in grape juice (must) into ethanol. Yeast is normally already present on grape skins. Fermentation can be done with this endogenous "wild yeast",[66] but this procedure gives unpredictable results, which depend upon the exact types of yeast species present. For this reason, a pure yeast culture is usually added to the must; this yeast quickly dominates the fermentation. The wild yeasts are repressed, which ensures a reliable and predictable fermentation.[67]
Most added wine yeasts are strains of S. cerevisiae, though not all strains of the species are suitable.[67] Different S. cerevisiae yeast strains have differing physiological and fermentative properties, therefore the actual strain of yeast selected can have a direct impact on the finished wine.[68] Significant research has been undertaken into the development of novel wine yeast strains that produce atypical flavour profiles or increased complexity in wines.[69][70]
The growth of some yeasts, such as Zygosaccharomyces and Brettanomyces, in wine can result in wine faults and subsequent spoilage.[71] Brettanomyces produces an array of metabolites when growing in wine, some of which are volatile phenolic compounds. Together, these compounds are often referred to as "Brettanomyces character", and are often described as "antiseptic" or "barnyard" type aromas. Brettanomyces is a significant contributor to wine faults within the wine industry.[72]
Researchers from the University of British Columbia, Canada, have found a new strain of yeast that has reduced amines. The amines in red wine and Chardonnay produce off-flavors and cause headaches and hypertension in some people. About 30% of people are sensitive to biogenic amines, such as histamines.[73]
Baking
Yeast, most commonly S. cerevisiae, is used in baking as a leavening agent, converting the fermentable sugars present in dough into carbon dioxide. This causes the dough to expand or rise as gas forms pockets or bubbles. When the dough is baked, the yeast dies and the air pockets "set", giving the baked product a soft and spongy texture. The use of potatoes, water from potato boiling, eggs, or sugar in a bread dough accelerates the growth of yeast. Most yeasts used in baking are of the same species common in alcoholic fermentation. In addition, Saccharomyces exiguus (also known as S. minor), a wild yeast found on plants, fruits, and grains, is occasionally used for baking. In breadmaking, the yeast initially respires aerobically, producing carbon dioxide and water. When the oxygen is depleted, fermentation begins, producing ethanol as a waste product; however, this evaporates during baking.[74]

It is not known when yeast was first used to bake bread. The first records that show this use came from Ancient Egypt.[8] Researchers speculate a mixture of flour meal and water was left longer than usual on a warm day and the yeasts that occur in natural contaminants of the flour caused it to ferment before baking. The resulting bread would have been lighter and tastier than the normal flat, hard cake.

Today, there are several retailers of baker's yeast; one of the earlier developments in North America is Fleischmann's Yeast, in 1868. During World War II, Fleischmann's developed a granulated active dry yeast which did not require refrigeration, had a longer shelf life than fresh yeast, and rose twice as fast. Baker's yeast is also sold as a fresh yeast compressed into a square "cake". This form perishes quickly, so must be used soon after production. A weak solution of water and sugar can be used to determine whether yeast is expired.[75] In the solution, active yeast will foam and bubble as it ferments the sugar into ethanol and carbon dioxide. Some recipes refer to this as proofing the yeast, as it "proves" (tests) the viability of the yeast before the other ingredients are added. When a sourdough starter is used, flour and water are added instead of sugar; this is referred to as proofing the sponge.
When yeast is used for making bread, it is mixed with flour, salt, and warm water or milk. The dough is kneaded until it is smooth, and then left to rise, sometimes until it has doubled in size. The dough is then shaped into loaves. Some bread doughs are knocked back after one rising and left to rise again (this is called dough proofing) and then baked. A longer rising time gives a better flavor, but the yeast can fail to raise the bread in the final stages if it is left for too long initially.
Bioremediation
Some yeasts can find potential application in the field of bioremediation. One such yeast, Yarrowia lipolytica, is known to degrade palm oil mill effluent, TNT (an explosive material), and other hydrocarbons, such as alkanes, fatty acids, fats and oils.[76] It can also tolerate high concentrations of salt and heavy metals,[77] and is being investigated for its potential as a heavy metal biosorbent.[78] Saccharomyces cerevisiae has potential to bioremediate toxic pollutants like arsenic from industrial effluent.[79] Bronze statues are known to be degraded by certain species of yeast.[80] Different yeasts from Brazilian gold mines bioaccumulate free and complexed silver ions.[81]
Industrial ethanol production
The ability of yeast to convert sugar into ethanol has been harnessed by the biotechnology industry to produce ethanol fuel. The process starts by milling a feedstock, such as sugar cane, field corn, or other cereal grains, and then adding dilute sulfuric acid, or fungal alpha amylase enzymes, to break down the starches into complex sugars. A glucoamylase is then added to break the complex sugars down into simple sugars. After this, yeasts are added to convert the simple sugars to ethanol, which is then distilled off to obtain ethanol up to 96% in purity.[82]
Saccharomyces yeasts have been genetically engineered to ferment xylose, one of the major fermentable sugars present in cellulosic biomasses, such as agriculture residues, paper wastes, and wood chips.[83][84] Such a development means ethanol can be efficiently produced from more inexpensive feedstocks, making cellulosic ethanol fuel a more competitively priced alternative to gasoline fuels.[85]
Nonalcoholic beverages


A number of sweet carbonated beverages can be produced by the same methods as beer, except the fermentation is stopped sooner, producing carbon dioxide, but only trace amounts of alcohol, leaving a significant amount of residual sugar in the drink.
- Root beer, originally made by Native Americans, commercialized in the United States by Charles Elmer Hires and especially popular during Prohibition
- Kvass, a fermented drink made from rye, popular in Eastern Europe. It has a recognizable, but low alcoholic content.[86]
- Kombucha, a fermented sweetened tea. Yeast in symbiosis with acetic acid bacteria is used in its preparation. Species of yeasts found in the tea can vary, and may include: Brettanomyces bruxellensis, Candida stellata, Schizosaccharomyces pombe, Torulaspora delbrueckii and Zygosaccharomyces bailii.[87] Also popular in Eastern Europe and some former Soviet republics under the name chajnyj grib (Russian: Чайный гриб), which means "tea mushroom".
- Kefir and kumis are made by fermenting milk with yeast and bacteria.[88]
- Mauby (Spanish: mabí), made by fermenting sugar with the wild yeasts naturally present on the bark of the Colubrina elliptica tree, popular in the Caribbean
Foods and Nutritional supplements


Yeast is used as an ingredient in foods for its umami flavor, in much of the same way that monosodium glutamate (MSG) is used and, like MSG, often contain free glutamic acid. Examples include:[89]
- Yeast extract, made from the intracellular contents of yeast and used as food additives or flavours. The general method for making yeast extract for food products such as Vegemite and Marmite on a commercial scale is heat autolysis, i.e. to add salt to a suspension of yeast, making the solution hypertonic, which leads to the cells' shrivelling up. This triggers autolysis, wherein the yeast's digestive enzymes break their own proteins down into simpler compounds, a process of self-destruction. The dying yeast cells are then heated to complete their breakdown, after which the husks (yeast with thick cell walls that would give poor texture) are removed. Yeast autolysates are used in Vegemite and Promite (Australia); Marmite (the United Kingdom); the unrelated Marmite (New Zealand); Vitam-R (Germany); and Cenovis (Switzerland).
- Nutritional yeast, which is whole dried, deactivated yeast cells, usually S. cerevisiae. Usually in the form of yellow flake or powder, its nutty and umami flavor makes it a vegan substitute for cheese powder.[90] Another popular use is as a topping for popcorn. It can also be used in mashed and fried potatoes, as well as in scrambled eggs. It comes in the form of flakes, or as a yellow powder similar in texture to cornmeal. In Australia, it is sometimes sold as "savoury yeast flakes".[91]
 Nutritional yeast flakes are yellow in colour
Nutritional yeast flakes are yellow in colour
Both types of yeast foods above are rich in B-complex vitamins (besides vitamin B12 unless fortified),[60] making them an attractive nutritional supplement to vegans.[90] The same vitamins are also found in some yeast-fermented products mentioned above, such as kvass.[92] Nutritional yeast in particular is naturally low in fat and sodium and a source of protein and vitamins as well as other minerals and cofactors required for growth. Many brands of nutritional yeast and yeast extract spreads, though not all, are fortified with vitamin B12, which is produced separately by bacteria.[93]
In 1920, the Fleischmann Yeast Company began to promote yeast cakes in a "Yeast for Health" campaign. They initially emphasized yeast as a source of vitamins, good for skin and digestion. Their later advertising claimed a much broader range of health benefits, and was censured as misleading by the Federal Trade Commission. The fad for yeast cakes lasted until the late 1930s.[94]
Probiotics
Some probiotic supplements use the yeast S. boulardii to maintain and restore the natural flora in the gastrointestinal tract. S. boulardii has been shown to reduce the symptoms of acute diarrhea,[95] reduce the chance of infection by Clostridium difficile (often identified simply as C. difficile or C. diff),[96] reduce bowel movements in diarrhea-predominant IBS patients,[97] and reduce the incidence of antibiotic-, traveler's-, and HIV/AIDS-associated diarrheas.[98]
Aquarium hobby
Yeast is often used by aquarium hobbyists to generate carbon dioxide (CO2) to nourish plants in planted aquaria.[99] CO2 levels from yeast are more difficult to regulate than those from pressurized CO2 systems. However, the low cost of yeast makes it a widely used alternative.[99]
Scientific research

Several yeasts, in particular S. cerevisiae and S. pombe, have been widely used in genetics and cell biology, largely because they are simple eukaryotic cells, serving as a model for all eukaryotes, including humans, for the study of fundamental cellular processes such as the cell cycle, DNA replication, recombination, cell division, and metabolism. Also, yeasts are easily manipulated and cultured in the laboratory, which has allowed for the development of powerful standard techniques, such as yeast two-hybrid,[100] synthetic genetic array analysis,[101] and tetrad analysis. Many proteins important in human biology were first discovered by studying their homologues in yeast; these proteins include cell cycle proteins, signaling proteins, and protein-processing enzymes.[102]
On 24 April 1996, S. cerevisiae was announced to be the first eukaryote to have its genome, consisting of 12 million base pairs, fully sequenced as part of the Genome Project.[103] At the time, it was the most complex organism to have its full genome sequenced, and the work of seven years and the involvement of more than 100 laboratories to accomplish.[104] The second yeast species to have its genome sequenced was Schizosaccharomyces pombe, which was completed in 2002.[105][106] It was the sixth eukaryotic genome sequenced and consists of 13.8 million base pairs. As of 2014, over 50 yeast species have had their genomes sequenced and published.[107]
Genomic and functional gene annotation of the two major yeast models can be accessed via their respective model organism databases: SGD[108][109] and PomBase.[110][111]
Genetically engineered biofactories
Various yeast species have been genetically engineered to efficiently produce various drugs, a technique called metabolic engineering.[112] S. cerevisiae is easy to genetically engineer; its physiology, metabolism and genetics are well known, and it is amenable for use in harsh industrial conditions. A wide variety of chemical in different classes can be produced by engineered yeast, including phenolics, isoprenoids, alkaloids, and polyketides.[113] About 20% of biopharmaceuticals are produced in S. cerevisiae, including insulin, vaccines for hepatitis, and human serum albumin.[114]
Pathogenic yeasts

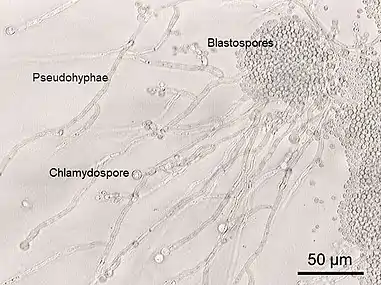
Some species of yeast are opportunistic pathogens that can cause infection in people with compromised immune systems. Cryptococcus neoformans and Cryptococcus gattii are significant pathogens of immunocompromised people. They are the species primarily responsible for cryptococcosis, a fungal infection that occurs in about one million HIV/AIDS patients, causing over 600,000 deaths annually.[115] The cells of these yeast are surrounded by a rigid polysaccharide capsule, which helps to prevent them from being recognised and engulfed by white blood cells in the human body.[116]
Yeasts of the genus Candida, another group of opportunistic pathogens, cause oral and vaginal infections in humans, known as candidiasis. Candida is commonly found as a commensal yeast in the mucous membranes of humans and other warm-blooded animals. However, sometimes these same strains can become pathogenic. The yeast cells sprout a hyphal outgrowth, which locally penetrates the mucosal membrane, causing irritation and shedding of the tissues.[117] A book from the 1980s listed the pathogenic yeasts of candidiasis in probable descending order of virulence for humans as: C. albicans, C. tropicalis, C. stellatoidea, C. glabrata, C. krusei, C. parapsilosis, C. guilliermondii, C. viswanathii, C. lusitaniae, and Rhodotorula mucilaginosa.[118] Candida glabrata is the second most common Candida pathogen after C. albicans, causing infections of the urogenital tract, and of the bloodstream (candidemia).[119] C. auris has been more recently identified.
Food spoilage
Yeasts are able to grow in foods with a low pH (5.0 or lower) and in the presence of sugars, organic acids, and other easily metabolized carbon sources.[120] During their growth, yeasts metabolize some food components and produce metabolic end products. This causes the physical, chemical, and sensible properties of a food to change, and the food is spoiled.[121] The growth of yeast within food products is often seen on their surfaces, as in cheeses or meats, or by the fermentation of sugars in beverages, such as juices, and semiliquid products, such as syrups and jams.[120] The yeast of the genus Zygosaccharomyces have had a long history as spoilage yeasts within the food industry. This is mainly because these species can grow in the presence of high sucrose, ethanol, acetic acid, sorbic acid, benzoic acid, and sulfur dioxide concentrations,[71] representing some of the commonly used food preservation methods. Methylene blue is used to test for the presence of live yeast cells.[122] In oenology, the major spoilage yeast is Brettanomyces bruxellensis.
Candida blankii has been detected in Iberian ham and meat.[123]
Symbiosis
An Indian study of seven bee species and nine plant species found 45 yeast species from 16 genera colonise the nectaries of flowers and honey stomachs of bees. Most were members of the genus Candida; the most common species in honey bee stomachs was Dekkera intermedia, while the most common species colonising flower nectaries was Candida blankii. Although the mechanics are not fully understood, it was found that A. indica flowers more if Candida blankii is present.[36]
In another example, Spathaspora passalidarum, found in the digestive tract of bess beetles, aids the digestion of plant cells by fermenting xylose.[124]
See also
- Bioaerosol
- Ethanol fermentation
- Evolution of aerobic fermentation
- Kazachstania yasuniensis – a yeast isolated in 2015
- Mycosis (fungal infection in animals)
- Start point (yeast)
- WHI3
- Yeast plasmids
- Zymology
References
- ↑ Piškur, Jure; Compagno, Concetta (2014). Molecular Mechanisms in Yeast Carbon Metabolism. Springer. p. 98. ISBN 978-3-642-55013-3.
The second completely sequenced yeast genome came 6 years later from the fission yeast Schizosaccharomyces pombe, which diverged from S. cerevisiae probably more than 300 million years ago.
- ↑ Kurtzman CP, Fell JW (2006). "Yeast Systematics and Phylogeny—Implications of Molecular Identification Methods for Studies in Ecology". Biodiversity and Ecophysiology of Yeasts, The Yeast Handbook. Springer.
- 1 2 Hoffman CS, Wood V, Fantes PA (October 2015). "An Ancient Yeast for Young Geneticists: A Primer on the Schizosaccharomyces pombe Model System". Genetics. 201 (2): 403–23. doi:10.1534/genetics.115.181503. PMC 4596657. PMID 26447128.
- ↑ Kurtzman CP, Piškur J (2006). "Taxonomy and phylogenetic diversity among the yeasts". In Sunnerhagen P, Piskur J (eds.). Comparative Genomics: Using Fungi as Models. Topics in Current Genetics. Vol. 15. Berlin: Springer. pp. 29–46. doi:10.1007/b106654. ISBN 978-3-540-31480-6.
- ↑ Kurtzman CP, Fell JW (2005). Gábor P, de la Rosa CL (eds.). Biodiversity and Ecophysiology of Yeasts. The Yeast Handbook. Berlin: Springer. pp. 11–30. ISBN 978-3-540-26100-1.
- ↑ Yong E (16 January 2012). "Yeast suggests speedy start for multicellular life". Nature. doi:10.1038/nature.2012.9810. S2CID 84392827.
- ↑ Walker K, Skelton H, Smith K (2002). "Cutaneous lesions showing giant yeast forms of Blastomyces dermatitidis". Journal of Cutaneous Pathology. 29 (10): 616–618. doi:10.1034/j.1600-0560.2002.291009.x. PMID 12453301. S2CID 39904013.
- 1 2 Legras JL, Merdinoglu D, Cornuet JM, Karst F (2007). "Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history". Molecular Ecology. 16 (10): 2091–2102. Bibcode:2007MolEc..16.2091L. doi:10.1111/j.1365-294X.2007.03266.x. PMID 17498234. S2CID 13157807.
- 1 2 Ostergaard S, Olsson L, Nielsen J (2000). "Metabolic Engineering of Saccharomyces cerevisiae". Microbiology and Molecular Biology Reviews. 64 (1): 34–50. doi:10.1128/MMBR.64.1.34-50.2000. PMC 98985. PMID 10704473.
- ↑ "Bioprocess automation". Helsinki University of Technology. 2007. Archived from the original on 7 May 2010. Retrieved 15 January 2012.
- ↑ Kurtzman CP (1994). "Molecular taxonomy of the yeasts". Yeast. 10 (13): 1727–1740. doi:10.1002/yea.320101306. PMID 7747515. S2CID 44797575.
- ↑ "What are yeasts?". Yeast Virtual Library. 13 September 2009. Archived from the original on 26 February 2009. Retrieved 28 November 2009.
- ↑ "Appendix I". Indo-European Roots (4th ed.). 2000. Archived from the original on 6 December 2008. Retrieved 16 November 2008.
{{cite book}}:|work=ignored (help) - 1 2 Phillips T. "Planets in a bottle: more about yeast". Science@NASA. Retrieved 3 October 2016.
- ↑ Aouizerat, Tzemach; Gutman, Itai; Paz, Yitzhak; Maeir, Aren M.; Gadot, Yuval; Gelman, Daniel; Szitenberg, Amir; Drori, Elyashiv; Pinkus, Ania; Schoemann, Miriam; Kaplan, Rachel; Ben-Gedalya, Tziona; Coppenhagen-Glazer, Shunit; Reich, Eli; Saragovi, Amijai; Lipschits, Oded; Klutstein, Michael; Hazan, Ronen (2019). "Isolation and Characterization of Live Yeast Cells from Ancient Vessels as a Tool in Bio-Archaeology". mBio. 10 (2). doi:10.1128/mBio.00388-19. PMC 6495373. PMID 31040238.
- ↑ Huxley A (1871). "Discourses: Biological & Geological (volume VIII) : Yeast". Collected Essays. Retrieved 28 November 2009.
- ↑ Ainsworth GC (1976). Introduction to the History of Mycology. Cambridge, UK: Cambridge University Press. p. 212. ISBN 9780521210133.
- ↑ Schwann T (1837). "Vorläufige Mittheilung, bettreffend Versuche über die Weingährung und Fäulniss". Annalen der Physik und Chemie (in German). 41 (5): 184–193. Bibcode:1837AnP...117..184S. doi:10.1002/andp.18371170517.
- ↑ Barnett JA (2004). "A history of research on yeasts 8: taxonomy". Yeast. 21 (14): 1141–1193. doi:10.1002/yea.1154. PMID 15515119. S2CID 34671745.
- ↑ Barnett JA (2003). "Beginnings of microbiology and biochemistry: the contribution of yeast research" (PDF). Microbiology. 149 (3): 557–567. doi:10.1099/mic.0.26089-0. PMID 12634325. S2CID 15986927. Archived from the original (PDF) on 3 March 2019.
- ↑ Klieger PC (2004). The Fleischmann yeast family. Arcadia Publishing. p. 13. ISBN 978-0-7385-3341-4.
- ↑ "Le Comité des Fabricants de levure". COFALEC. Archived from the original on 14 May 2010. Retrieved 21 February 2010.
- ↑ Snodgrass ME (2004). Encyclopedia of Kitchen History. New York, New York: Fitzroy Dearborn. p. 1066. ISBN 978-1-57958-380-4.
- ↑ Denison, Merrill (1955). The Barley and the Stream: The Molson Story. Toronto: McClelland & Stewart Limited. p. 165.
- ↑ Barnett JA (1975). "The entry of D-ribose into some yeasts of the genus Pichia". Journal of General Microbiology. 90 (1): 1–12. doi:10.1099/00221287-90-1-1. PMID 1176959.
- ↑ Arthur H, Watson K (1976). "Thermal adaptation in yeast: growth temperatures, membrane lipid, and cytochrome composition of psychrophilic, mesophilic, and thermophilic yeasts". Journal of Bacteriology. 128 (1): 56–68. doi:10.1128/JB.128.1.56-68.1976. PMC 232826. PMID 988016.
- ↑ Kaufmann K, Schoneck A (2002). Making Sauerkraut and Pickled Vegetables at Home: Creative Recipes for Lactic Fermented Food to Improve Your Health. Book Publishing Company. ISBN 978-1-55312-037-7.
- ↑ Suh SO, McHugh JV, Pollock DD, Blackwell M (2005). "The beetle gut: a hyperdiverse source of novel yeasts". Mycological Research. 109 (3): 261–265. doi:10.1017/S0953756205002388. PMC 2943959. PMID 15912941.
- ↑ Sláviková E, Vadkertiová R (2003). "The diversity of yeasts in the agricultural soil". Journal of Basic Microbiology. 43 (5): 430–436. doi:10.1002/jobm.200310277. PMID 12964187. S2CID 12030027.
- ↑ Martin, Phillip L.; King, William; Bell, Terrence H; Peter, Kari (2021). "The decay and fungal succession of apples with bitter rot across a vegetation diversity gradient". Phytobiomes Journal. 6: 26–34. doi:10.1094/pbiomes-06-21-0039-r. ISSN 2471-2906. S2CID 239658496.
- 1 2 Herrera C, Pozo MI (2010). "Nectar yeasts warm the flowers of a winter-blooming plant". Proceedings of the Royal Society B. 277 (1689): 1827–1834. doi:10.1098/rspb.2009.2252. PMC 2871880. PMID 20147331.
- ↑ Oyeka CA, Ugwu LO (2002). "Fungal flora of human toe webs". Mycoses. 45 (11–12): 488–491. doi:10.1046/j.1439-0507.2002.00796.x. PMID 12472726. S2CID 8789635.
- ↑ Martini A (1992). "Biodiversity and conservation of yeasts". Biodiversity and Conservation. 1 (4): 324–333. Bibcode:1992BiCon...1..324M. doi:10.1007/BF00693768. S2CID 35231385.
- ↑ Bass D, Howe A, Brown N, Barton H, Demidova M, Michelle H, Li L, Sanders H, Watkinson SC, Willcock S, Richards TA (2007). "Yeast forms dominate fungal diversity in the deep oceans". Proceedings of the Royal Society B. 274 (1629): 3069–3077. doi:10.1098/rspb.2007.1067. PMC 2293941. PMID 17939990.
- ↑ Kutty SN, Philip R (2008). "Marine yeasts—a review" (PDF). Yeast. 25 (7): 465–483. doi:10.1002/yea.1599. PMID 18615863. S2CID 26625932.
- 1 2 Sandhu DK, Waraich MK (1985). "Yeasts associated with pollinating bees and flower nectar". Microbial Ecology. 11 (1): 51–58. Bibcode:1985MicEc..11...51S. doi:10.1007/BF02015108. JSTOR 4250820. PMID 24221239. S2CID 1776642.
- ↑ Barley S (10 February 2010). "Stinky flower is kept warm by yeast partner". New Scientist. (subscription required)
- ↑ Little AEF, Currie CR (2008). "Black yeast symbionts compromise the efficiency of antibiotic defenses in fungus-growing ants". Ecology. 89 (5): 1216–1222. Bibcode:2008Ecol...89.1216L. doi:10.1890/07-0815.1. PMID 18543616. S2CID 28969854.
- ↑ Magliani W, Conti S, Frazzi R, Ravanetti L, Maffei DL, Polonelli L (2006). "Protective antifungal yeast killer toxin-like antibodies". Current Molecular Medicine. 5 (4): 443–452. doi:10.2174/1566524054022558. PMID 15978000.
- 1 2 3 Zaky, Abdelrahman Saleh; Tucker, Gregory A.; Daw, Zakaria Yehia; Du, Chenyu (September 2014). "Marine yeast isolation and industrial application". FEMS Yeast Research. 14 (6): 813–825. doi:10.1111/1567-1364.12158. PMC 4262001. PMID 24738708.
 This article contains quotations from this source, which is available under a Creative Commons Attribution license.
This article contains quotations from this source, which is available under a Creative Commons Attribution license. - 1 2 Kutty, Sreedevi N.; Philip, Rosamma (July 2008). "Marine yeasts—a review". Yeast. 25 (7): 465–483. doi:10.1002/yea.1599. PMID 18615863. S2CID 26625932.
- ↑ Zaky, Abdelrahman Saleh; Greetham, Darren; Louis, Edward J.; Tucker, Greg A.; Du, Chenyu (28 November 2016). "A New Isolation and Evaluation Method for Marine-Derived Yeast spp. with Potential Applications in Industrial Biotechnology". Journal of Microbiology and Biotechnology. 26 (11): 1891–1907. doi:10.4014/jmb.1605.05074. PMID 27435537. S2CID 40476719.
- ↑ Zaky, Abdelrahman Saleh; Greetham, Darren; Tucker, Gregory A.; Du, Chenyu (14 August 2018). "The establishment of a marine focused biorefinery for bioethanol production using seawater and a novel marine yeast strain". Scientific Reports. 8 (1): 12127. Bibcode:2018NatSR...812127Z. doi:10.1038/s41598-018-30660-x. ISSN 2045-2322. PMC 6092365. PMID 30108287.
- 1 2 Balasubramanian MK, Bi E, Glotzer M (2004). "Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells". Current Biology. 14 (18): R806–818. doi:10.1016/j.cub.2004.09.022. PMID 15380095. S2CID 12808612.
- ↑ Yeong FM (2005). "Severing all ties between mother and daughter: cell separation in budding yeast". Molecular Microbiology. 55 (5): 1325–1331. doi:10.1111/j.1365-2958.2005.04507.x. PMID 15720543. S2CID 25013111.
- ↑ Neiman AM (2005). "Ascospore formation in the yeast Saccharomyces cerevisiae". Microbiology and Molecular Biology Reviews. 69 (4): 565–584. doi:10.1128/MMBR.69.4.565-584.2005. PMC 1306807. PMID 16339736.
- ↑ Davey J (1998). "Fusion of a fission yeast". Yeast. 14 (16): 1529–1566. doi:10.1002/(SICI)1097-0061(199812)14:16<1529::AID-YEA357>3.0.CO;2-0. PMID 9885154. S2CID 44652765.
- ↑ Bernstein C, Johns V (1989). "Sexual reproduction as a response to H2O2 damage in Schizosaccharomyces pombe". Journal of Bacteriology. 171 (4): 1893–1897. doi:10.1128/jb.171.4.1893-1897.1989. PMC 209837. PMID 2703462.
- ↑ Herskowitz I (1988). "Life cycle of the budding yeast Saccharomyces cerevisiae". Microbiological Reviews. 52 (4): 536–553. doi:10.1128/MMBR.52.4.536-553.1988. PMC 373162. PMID 3070323.
- ↑ Katz Ezov T, Chang SL, Frenkel Z, Segrè AV, Bahalul M, Murray AW, Leu JY, Korol A, Kashi Y (2010). "Heterothallism in Saccharomyces cerevisiae isolates from nature: effect of HO locus on the mode of reproduction". Molecular Ecology. 19 (1): 121–131. Bibcode:2010MolEc..19..121K. doi:10.1111/j.1365-294X.2009.04436.x. PMC 3892377. PMID 20002587.
- 1 2 Ruderfer DM, Pratt SC, Seidel HS, Kruglyak L (2006). "Population genomic analysis of outcrossing and recombination in yeast". Nature Genetics. 38 (9): 1077–1081. doi:10.1038/ng1859. PMID 16892060. S2CID 783720.
- ↑ Birdsell JA, Wills C (2003). MacIntyre RJ, Clegg MT (eds.). The evolutionary origin and maintenance of sexual recombination: A review of contemporary models. Evolutionary Biology Series >> Evolutionary Biology. Vol. 33. Springer. pp. 27–137. ISBN 978-0306472619.
- ↑ Bai FY, Zhao JH, Takashima M, Jia JH, Boekhout T, Nakase T (2002). "Reclassification of the Sporobolomyces roseus and Sporidiobolus pararoseus complexes, with the description of Sporobolomyces phaffii sp. nov". International Journal of Systematic and Evolutionary Microbiology. 52 (6): 2309–2314. doi:10.1099/ijs.0.02297-0. PMID 12508902.
- ↑ Chen X, Jiang ZH, Chen S, Qin W (2010). "Microbial and bioconversion production of D-xylitol and its detection and application". International Journal of Biological Sciences. 6 (7): 834–844. doi:10.7150/ijbs.6.834. PMC 3005349. PMID 21179590.

- ↑ Botstein D, Fink GR (2011). "Yeast: an experimental organism for 21st Century biology". Genetics. 189 (3): 695–704. doi:10.1534/genetics.111.130765. PMC 3213361. PMID 22084421.

- ↑ Priest FG, Stewart GG (2006). Handbook of Brewing. CRC Press. p. 84. ISBN 9781420015171.
- ↑ Gibson M (2010). The Sommelier Prep Course: An Introduction to the Wines, Beers, and Spirits of the World. John Wiley and Sons. p. 361. ISBN 978-0-470-28318-9.
- ↑ For more on the taxonomical differences, see Dowhanick TM (1999). "Yeast – Strains and Handling Techniques". In McCabe JT (ed.). The Practical Brewer. Master Brewers Association of the Americas.
- ↑ Amendola J, Rees N (2002). Understanding Baking: The Art and Science of Baking. John Wiley and Sons. p. 36. ISBN 978-0-471-40546-7.
- 1 2 "Brewer's yeast". University of Maryland Medical Center. Archived from the original on 2 July 2017.
- ↑ Vanderhaegen B, Neven H, Cogne S, Vertrepin KJ, Derdelinckx C, Verachtert H (2003). "Bioflavoring and Beer Refermentation". Applied Microbiology and Biotechnology. 62 (2–3): 140–150. doi:10.1007/s00253-003-1340-5. PMID 12759790. S2CID 12944068.
- ↑ Custers MTJ (1940). Onderzoekingen over het gistgeslacht Brettanomyces (PhD thesis) (in Dutch). Delft, the Netherlands: Delft University.
- ↑ Van der Walt JP (1984). "The Yeasts: A Taxonomic Study" (3rd ed.). Amsterdam: Elsevier Science: 146–150.
{{cite journal}}: Cite journal requires|journal=(help) - ↑ Oelofse A, Pretorius IS, du Toit M (2008). "Significance of Brettanomyces and Dekkera during winemaking: a synoptic review" (PDF). South African Journal of Enology and Viticulture. 29 (2): 128–144.
- ↑ Yakobson CM (2010). Pure culture fermentation characteristics of Brettanomyces yeast species and their use in the brewing industry (MSc.). International Centre for Brewing and Distilling, Heriot-Watt University.
- ↑ Ross JP (September 1997). "Going wild: wild yeast in winemaking". Wines & Vines. Archived from the original on 5 May 2005. Retrieved 15 January 2012.
- 1 2 González Techera A, Jubany S, Carrau FM, Gaggero C (2001). "Differentiation of industrial wine yeast strains using microsatellite markers". Letters in Applied Microbiology. 33 (1): 71–75. doi:10.1046/j.1472-765X.2001.00946.x. PMID 11442819. S2CID 7625171.
- ↑ Dunn B, Levine RP, Sherlock G (2005). "Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures". BMC Genomics. 6 (1): 53. doi:10.1186/1471-2164-6-53. PMC 1097725. PMID 15833139.
- ↑ "Research enables yeast supplier to expands options" (PDF). Archived from the original (PDF) on 21 September 2006. Retrieved 10 January 2007.
- ↑ McBryde C, Gardner JM, de Barros Lopes M, Jiranek V (2006). "Generation of novel wine yeast strains by adaptive evolution". American Journal of Enology and Viticulture. 57 (4): 423–430. doi:10.5344/ajev.2006.57.4.423. S2CID 83723719.
- 1 2 Loureiro V, Malfeito-Ferreira M (2003). "Spoilage yeasts in the wine industry". International Journal of Food Microbiology. 86 (1–2): 23–50. doi:10.1016/S0168-1605(03)00246-0. PMID 12892920.
- ↑ Lamar J. "Brettanomyces (Dekkera)". Vincyclopedia. Retrieved 28 November 2009.
- ↑ Shore R (15 February 2011). "Eureka! Vancouver scientists take the headache out of red wine". The Vancouver Sun. Archived from the original on 17 February 2011.
- ↑ Moore-Landecker E (1996). Fundamentals of the Fungi. Englewood Cliffs, NJ: Prentice Hall. pp. 533–534. ISBN 978-0-13-376864-0.
- ↑ John (24 August 2023). "Does Yeast Expire? [Active Dry vs Instant Yeast]". PizzaOvensHub. Retrieved 27 September 2023.
- ↑ Zinjarde S, Apte M, Mohite P, Kumar AR (2014). "Yarrowia lipolytica and pollutants: Interactions and applications". Biotechnology Advances. 32 (5): 920–933. doi:10.1016/j.biotechadv.2014.04.008. PMID 24780156.
- ↑ Bankar AV, Kumar AR, Zinjarde SS (2009). "Environmental and industrial applications of Yarrowia lipolytica". Applied Microbiology and Biotechnology. 84 (5): 847–865. doi:10.1007/s00253-009-2156-8. PMID 19669134. S2CID 38670765.
- ↑ Bankar AV, Kumar AR, Zinjarde SS (2009). "Removal of chromium (VI) ions from aqueous solution by adsorption onto two marine isolates of Yarrowia lipolytica". Journal of Hazardous Materials. 170 (1): 487–494. doi:10.1016/j.jhazmat.2009.04.070. PMID 19467781.
- ↑ Soares EV, Soares HMVM (2012). "Bioremediation of industrial effluents containing heavy metals using brewing cells of Saccharomyces cerevisiae as a green technology: A review" (PDF). Environmental Science and Pollution Research. 19 (4): 1066–1083. Bibcode:2012ESPR...19.1066S. doi:10.1007/s11356-011-0671-5. hdl:10400.22/10260. PMID 22139299. S2CID 24030739.
- ↑ Cappitelli F, Sorlini C (2008). "Microorganisms attack synthetic polymers in items representing our cultural heritage". Applied and Environmental Microbiology. 74 (3): 564–569. Bibcode:2008ApEnM..74..564C. doi:10.1128/AEM.01768-07. PMC 2227722. PMID 18065627.

- ↑ Singh H (2006). Mycoremediation: Fungal Bioremediation. John Wiley & Sons. p. 507. ISBN 978-0-470-05058-3.
- ↑ "Fuel Ethanol Production: GSP Systems Biology Research". Genomic Science Program. U.S. Department of Energy Office of Science. Archived from the original on 3 June 2009. Retrieved 28 November 2009.
- ↑ Brat D, Boles E, Wiedemann B (2009). "Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae". Applied and Environmental Microbiology. 75 (8): 2304–2311. Bibcode:2009ApEnM..75.2304B. doi:10.1128/AEM.02522-08. PMC 2675233. PMID 19218403.
- ↑ Ho NW, Chen Z, Brainard AP (1998). "Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose". Applied and Environmental Microbiology. 64 (5): 1852–1859. Bibcode:1998ApEnM..64.1852H. doi:10.1128/AEM.64.5.1852-1859.1998. PMC 106241. PMID 9572962.
- ↑ Madhavan A, Srivastava A, Kondo A, Bisaria VS (2012). "Bioconversion of lignocellulose-derived sugars to ethanol by engineered Saccharomyces cerevisiae". Critical Reviews in Biotechnology. 32 (1): 22–48. doi:10.3109/07388551.2010.539551. PMID 21204601. S2CID 207467678.
- ↑ Smith A, Kraig B (2013). The Oxford Encyclopedia of Food and Drink in America. Oxford University Press. p. 440. ISBN 978-0-19-973496-2.
- ↑ Teoh AL, Heard G, Cox J (2004). "Yeast ecology of Kombucha fermentation". International Journal of Food Microbiology. 95 (2): 119–126. doi:10.1016/j.ijfoodmicro.2003.12.020. PMID 15282124.
- ↑ de Oliveira Leite AM, Miguel MA, Peixoto RS, Rosado AS, Silva JT, Paschoalin VM (2013). "Microbiological, technological and therapeutic properties of kefir: A natural probiotic beverage". Brazilian Journal of Microbiology. 44 (2): 341–349. doi:10.1590/S1517-83822013000200001. PMC 3833126. PMID 24294220.

- ↑ Stewart, Graham G.; Priest, Fergus G. (22 February 2006). Handbook of Brewing, Second Edition. CRC Press. p. 691. ISBN 978-1-4200-1517-1.
- 1 2 Thaler M, Safferstein D (2014). A Curious Harvest: The Practical Art of Cooking Everything. Quarry Books. p. 129. ISBN 978-1-59253-928-4.
- ↑ Lee JG (ed.). "South East Asia Under Japanese Occupation – Harukoe (Haruku)". Children (& Families) of the Far East Prisoners of War. Retrieved 28 November 2009.
- ↑ Līdums, Ivo; Kārkliņa, Daina; Ķirse, Asnate; Šabovics, Mārtiņš (April 2017). "Nutritional value, vitamins, sugars and aroma volatiles in naturally fermented and dry kvass" (PDF). Foodbalt. Faculty of Food Technology, Latvia University of Life Sciences and Technologies: 61–65. doi:10.22616/foodbalt.2017.027. ISSN 2501-0190.
- ↑ Duyff RL (2012). American Dietetic Association Complete Food and Nutrition Guide, Revised and Updated (4th ed.). Houghton Mifflin Harcourt. pp. 256–257. ISBN 978-0-544-66456-2.
- ↑ Price C (Fall 2015). "The healing power of compressed yeast". Distillations Magazine. 1 (3): 17–23. Retrieved 20 March 2018.
- ↑ Dinleyici EC, Eren M, Ozen M, Yargic ZA, Vandenplas Y (2012). "Effectiveness and safety of Saccharomyces boulardii for acute infectious diarrhea". Expert Opinion on Biological Therapy. 12 (4): 395–410. doi:10.1517/14712598.2012.664129. PMID 22335323. S2CID 40040866.
- ↑ Johnson S, Maziade PJ, McFarland LV, Trick W, Donskey C, Currie B, Low DE, Goldstein EJ (2012). "Is primary prevention of Clostridium difficile infection possible with specific probiotics?". International Journal of Infectious Diseases. 16 (11): e786–92. doi:10.1016/j.ijid.2012.06.005. PMID 22863358.
- ↑ Dai C, Zheng CQ, Jiang M, Ma XY, Jiang LJ (2013). "Probiotics and irritable bowel syndrome". World Journal of Gastroenterology. 19 (36): 5973–5980. doi:10.3748/wjg.v19.i36.5973. PMC 3785618. PMID 24106397.

- ↑ McFarland LV (2010). "Systematic review and meta-analysis of Saccharomyces boulardii in adult patients". World Journal of Gastroenterology. 16 (18): 2202–22. doi:10.3748/wjg.v16.i18.2202. PMC 2868213. PMID 20458757.

- 1 2 Pedersen O, Andersen T, Christensen C (2007). "CO2 in planted aquaria" (PDF). The Aquatic Gardener. 20 (3): 24–33. Archived from the original (PDF) on 24 June 2016. Retrieved 29 May 2016.
- ↑ Brückner A, Polge C, Lentze N, Auerbach D, Schlattner U (2009). "Yeast two-hybrid, a powerful tool for systems biology". International Journal of Molecular Sciences. 10 (6): 2763–2788. doi:10.3390/ijms10062763. PMC 2705515. PMID 19582228.

- ↑ Tong AHY, Boone C (2006). "Synthetic genetic array analysis in Saccharomyces cerevisiae". In Xiao W. (ed.). Yeast Protocols. Springer Science & Business Media. pp. 171–191. ISBN 978-1-59259-958-5.
- ↑ Ishiwata S, Kuno T, Takada H, Koike A, Sugiura R (2007). "Molecular genetic approach to identify inhibitors of signal transduction pathways". In Conn PM (ed.). Sourcebook of Models for Biomedical Research. Springer Science & Business Media. pp. 439–444. ISBN 978-1-58829-933-8.
- ↑ Williams N (1996). "Genome Projects: Yeast genome sequence ferments new research". Science. 272 (5261): 481. Bibcode:1996Sci...272..481W. doi:10.1126/science.272.5261.481. PMID 8614793. S2CID 35565404.
- ↑ Henahan S (24 April 1996). "Complete DNA Sequence of Yeast". Science Updates. Archived from the original on 5 March 2012. Retrieved 15 January 2012.
- ↑ Wood V, Gwilliam R, Rajandream MA, et al. (2002). "The genome sequence of Schizosaccharomyces pombe" (PDF). Nature. 415 (6874): 871–880. doi:10.1038/nature724. PMID 11859360. S2CID 4393190.
- ↑ Reinert B (1 March 2002). "Schizosaccharomyces pombe: Second yeast genome sequenced". Genome News Network. Archived from the original on 3 May 2008. Retrieved 15 January 2012.
- ↑ Lin Z, Li W-H (2014). "Comparative genomics and evolutionary genetics of yeast carbon metabolism". In Piskur J, Compagno C (eds.). Molecular Mechanisms in Yeast Carbon Metabolism. Springer. p. 98. ISBN 978-3-642-55013-3.
- ↑ "About SGD". Saccharomyces Genome Database.
- ↑ Cherry, JM; Hong, EL; Amundsen, C; Balakrishnan, R; Binkley, G; Chan, ET; Christie, KR; Costanzo, MC; Dwight, SS; Engel, SR; Fisk, DG; Hirschman, JE; Hitz, BC; Karra, K; Krieger, CJ; Miyasato, SR; Nash, RS; Park, J; Skrzypek, MS; Simison, M; Weng, S; Wong, ED (January 2012). "Saccharomyces Genome Database: the genomics resource of budding yeast". Nucleic Acids Research. 40 (Database issue): D700–5. doi:10.1093/nar/gkr1029. PMC 3245034. PMID 22110037.
- ↑ "PomBase".
- ↑ Lock, A; Rutherford, K; Harris, MA; Hayles, J; Oliver, SG; Bähler, J; Wood, V (13 October 2018). "PomBase 2018: user-driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information". Nucleic Acids Research. 47 (D1): D821–D827. doi:10.1093/nar/gky961. PMC 6324063. PMID 30321395.
- ↑ N. Milne, P. Thomsen, N. Mølgaard Knudsen, P. Rubaszka, M. Kristensen, L. Borodina (1 July 2020). "Metabolic engineering of Saccharomyces cerevisiae for the de novo production of psilocybin and related tryptamine derivatives". Metabolic Engineering. 60: 25–36. doi:10.1016/j.ymben.2019.12.007. ISSN 1096-7176. PMC 7232020. PMID 32224264.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Siddiqui MS, Thodey K, Trenchard I, Smolke CD (2012). "Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools". FEMS Yeast Research. 12 (2): 144–170. doi:10.1111/j.1567-1364.2011.00774.x. PMID 22136110.

- ↑ Nilesen J (2012). "Production of biopharmaceutical proteins by yeast. Advances through metabolic engineering". Bioengineered. 4 (4): 207–211. doi:10.4161/bioe.22856. PMC 3728191. PMID 23147168.

- ↑ Cogliati M (2013). "Global molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii: An atlas of the molecular types". Scientifica. 2013: 675213. doi:10.1155/2013/675213. PMC 3820360. PMID 24278784.
- ↑ O'Meara TR, Alspaugh JA (2012). "The Cryptococcus neoformans capsule: A sword and a shield". Clinical Microbiology Reviews. 25 (3): 387–408. doi:10.1128/CMR.00001-12. PMC 3416491. PMID 22763631.

- ↑ Deacon J. "The Microbial World: Yeasts and yeast-like fungi". Institute of Cell and Molecular Biology. Archived from the original on 25 September 2006. Retrieved 18 September 2008.
- ↑ Hurley R, de Louvois J, Mulhall A (1987). "Yeast as human and animal pathogens". In Rose AH, Harrison JS (eds.). The Yeasts. Volume 1: Biology of Yeasts (2nd ed.). New York, New York: Academic Press. pp. 207–281.
- ↑ Brunke S, Hube B (2013). "Two unlike cousins: Candida albicans and C. glabrata infection strategies". Cellular Microbiology. 15 (5): 701–708. doi:10.1111/cmi.12091. PMC 3654559. PMID 23253282.

- 1 2 Kurtzman CP (2006). "Detection, identification and enumeration methods for spoilage yeasts". In Blackburn CDW (ed.). Food spoilage microorganisms. Cambridge, England: Woodhead Publishing. pp. 28–54. ISBN 978-1-85573-966-6.
- ↑ Fleet GH, Praphailong W (2001). "Yeasts". In Moir CJ (ed.). Spoilage of Processed Foods: Causes and Diagnosis. Food Microbiology Group of the Australian Institute of Food Science and Technology (AIFST). pp. 383–397. ISBN 978-0-9578907-0-1.
- ↑ Downes FP, Ito K (2001). Compendium of Methods for the Microbiological Examination of Foods. Washington, DC: American Public Health Association. p. 211. ISBN 978-0-87553-175-5.
- ↑ Toldrá, Fidel (October 2014). Toldrá, Fidel; Hui, Y. H.; Astiasaran, Iciar; Sebranek, Joseph; Talon, Regine (eds.). Handbook of Fermented Meat and Poultry (2nd ed.). Chichester, West Sussex, UK: Wiley-Blackwell. p. 140. ISBN 978-1-118-52267-7.
- ↑ Nguyen, Nhu H.; Suh, Sung-Oui; Marshall, Christopher J.; Blackwell, Meredith (1 October 2006). "Morphological and ecological similarities: wood-boring beetles associated with novel xylose-fermenting yeasts, Spathaspora passalidarum gen. sp. nov. and Candida jeffriesii sp. nov". Mycological Research. 110 (10): 1232–1241. doi:10.1016/j.mycres.2006.07.002. ISSN 0953-7562. PMID 17011177.
Further reading
- Alexopoulos CJ, Mims CW, Blackwell M (1996). Introductory Mycology. New York: Wiley. ISBN 978-0-471-52229-4.
- Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008). Dictionary of the Fungi (10th ed.). Wallingford, UK: CAB International. ISBN 978-0-85199-826-8.
- Kurtzman CP; Fell JW; Boekhout T, eds. (2011). The Yeasts: A Taxonomic Study. Vol. 1 (5th ed.). Amsterdam, etc.: Elsevier. ISBN 978-0-12-384708-9.
- Money, Nicholas P. (2018). The Rise of Yeast: How the Sugar Fungus Shaped Civilisation. Oxford University Press. ISBN 978-0198749707.
- Priest FG, Stewart GG (2006). Handbook of Brewing (2nd ed.). CRC Press. p. 691. ISBN 978-1-4200-1517-1.
External links
- Saccharomyces genome database
- Yeast growth and the cell cycle (archived 21 July 2007)
- Yeast virtual library
