| Neuraminidase | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
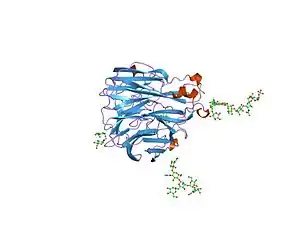
 Crystallographic structure of influenza neuraminidase in complex with the inhibitor 4-acetamido-3-hydroxy-5-nitro-benzoic acid.[1] | |||||||||||
| Identifiers | |||||||||||
| Symbol | Neur | ||||||||||
| Pfam | PF00064 | ||||||||||
| Pfam clan | CL0434 | ||||||||||
| InterPro | IPR001860 | ||||||||||
| SCOP2 | 2bat / SCOPe / SUPFAM | ||||||||||
| CAZy | GH34 | ||||||||||
| CDD | cd00260 | ||||||||||
| |||||||||||

Viral neuraminidase is a type of neuraminidase found on the surface of influenza viruses that enables the virus to be released from the host cell. Neuraminidases are enzymes that cleave sialic acid (also called neuraminic acid) groups from glycoproteins. Viral neuraminidase was discovered by Alfred Gottschalk at the Walter and Eliza Hall Institute in 1957.[3] Neuraminidase inhibitors are antiviral agents that inhibit influenza viral neuraminidase activity and are of major importance in the control of influenza.[4]
Viral neuraminidases are the members of the glycoside hydrolase family 34 CAZY GH_34 which comprises enzymes with only one known activity; sialidase or neuraminidase EC 3.2.1.18. Neuraminidases cleave the terminal sialic acid residues from carbohydrate chains in glycoproteins. Sialic acid is a negatively charged sugar associated with the protein and lipid portions of lipoproteins.
To infect a host cell, the influenza virus attaches to the exterior cell surface using hemagglutinin, a molecule found on the surface of the virus that binds to sialic acid groups. Sialic acids are found on various glycoproteins at the host cell surface. The virus then moves from sialic acid group to sialic acid group until it finds the proper cell surface receptor (whose identity remains unknown).[5] Neuraminadase enables this movement by cleaving sialic acid groups that hemagglutinin was attached to. After the virus has entered the cell and has replicated, new viral particles bud from the host cell membrane. The hemagglutinin on new viral particles remains attached to sialic acid groups of glycoproteins on the external cell surface and the surface of other viral particles; neuraminadase cleaves these groups and thereby allows the release of viral particles[6] and prevents self-aggregation.[5] Neuraminadase also facilitates the movement of virus particles in the presence of mucus rich in silicic acid.[5]
A single hemagglutinin-neuraminidase protein can combine neuraminidase and hemagglutinin functions, such as in mumps virus and human parainfluenza virus.
Function
The enzyme helps viruses to be released after budding from the plasma membrane of a host cell. Influenza virus membranes contain two glycoproteins: hemagglutinin and neuraminidase. While the hemagglutinin on the surface of the virion is needed for infection, its presence inhibits release of the particle after budding. Viral neuraminidase cleaves terminal sialic acid residues from glycan structures on the surface of the infected cell. This promotes the release of progeny viruses and the spread of the virus from the host cell to uninfected surrounding cells. Neuraminidase also cleaves sialic acid residues from viral proteins, preventing aggregation of viruses.
Inhibitors
Neuraminidase has been targeted in structure-based enzyme inhibitor design programmes that have resulted in the production of two drugs, zanamivir (Relenza) and oseltamivir (Tamiflu). Administration of neuraminidase inhibitors is a treatment that limits the severity and spread of viral infections. Neuraminidase inhibitors are useful for combating influenza infection: zanamivir, administered by inhalation; oseltamivir, administered orally; and under research is peramivir administered parenterally, that is through intravenous or intramuscular injection.
Neuraminidase inhibition resistance
On February 27, 2005, a 14-year-old Vietnamese girl was documented to be carrying an H5N1 influenza virus strain that was resistant to the drug oseltamivir. The drug is used to treat patients that have contracted influenza. However, the Vietnamese girl who had received a prophylactic dose (75 mg once a day) was found to be non-responsive to the medication. In growing fears of a global avian flu pandemic, scientists began to look for a cause of resistance to the Tamiflu medication. The cause was determined to be a histidine-to-tyrosine (amino acid) substitution at position 274 in its neuraminidase protein.
As strains of influenza are continuously mutating, it is essential that scientists quickly and efficiently determine the correct neuraminidase subtype that is responsible for the drug resistance in order to develop medications that will combat specific strains of influenza.
A new class of neuraminidase inhibitors that covalently attach to the enzyme have shown activity against drug-resistant virus in vitro.[7][8]
Specificity
In ideal circumstances, influenza virus neuraminidase (NA) should act on the same type of receptor the virus hemagglutinin (HA) binds to, a phenomenon that does not always happen. It is not quite clear how the virus manages to function when there is no close match between the specificities of NA and HA.
Exo- and endo-
Neuraminidase enzymes can have endo- or exo-glycosidase activity, and are classified as EC 3.2.1.29 (endo-neuraminidase)[9] and EC 3.2.1.18 (exo-neuraminidases).[10] In general, mammalian sialic acid residues are at terminal positions (non-reducing end) in complex glycans, and so viral neuraminidases - which are exo-glycosidase enzymes - use these terminal residues as their substrates.
See also
References
- ↑ Jedrzejas, MJ; Singh, S; Brouillette, WJ; Laver, WG; Air, GM; Luo, M (14 March 1995). "Structures of aromatic inhibitors of influenza virus neuraminidase". Biochemistry. 34 (10): 3144–51. doi:10.1021/bi00010a003. PMID 7880809.
- ↑ Varghese, J. N.; McKimm-Breschkin, J. L.; Caldwell, J. B.; Kortt, A. A.; Colman, P. M. (1992). "The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor". Proteins: Structure, Function, and Genetics. 14 (3): 327–32. doi:10.1002/prot.340140302. PMID 1438172. S2CID 41743465.
- ↑ "WEHI History: 1957 Discovery of Neuraminidase - Key Flu Molecule". WEHI. Retrieved 2023-11-08.
- ↑ Couch RB (1999). "Measures for control of influenza". PharmacoEconomics. 16 (Suppl 1): 41–5. doi:10.2165/00019053-199916001-00006. PMID 10623375. S2CID 41844816.
- 1 2 3 Dou, Dan; Revol, Rebecca; Östbye, Henrik; Wang, Hao; Daniels, Robert (2018-07-20). "Influenza A Virus Cell Entry, Replication, Virion Assembly and Movement". Frontiers in Immunology. 9: 1581. doi:10.3389/fimmu.2018.01581. ISSN 1664-3224. PMC 6062596. PMID 30079062.
- ↑ Huang IC, Li W, Sui J, Marasco W, Choe H, Farzan M (May 2008). "Influenza A virus neuraminidase limits viral superinfection". J. Virol. 82 (10): 4834–43. doi:10.1128/JVI.00079-08. PMC 2346733. PMID 18321971.
- ↑ BBC News: Flu drug 'shows promise' in overcoming resistance (accessed 22 February 2013)
- ↑ Kim J-H et al. Mechanism-based covalent neuraminidase inhibitors with broad spectrum influenza antiviral activity. Science doi:10.1126/science.1232552
- ↑ "EC 3.2.1.129".
- ↑ "EC 3.2.1.18".
External links
- Influenza Research Database Database of influenza sequences (including neuraminidase).
- Proteopedia Influenza Neuraminidase, Tamiflu and Relenza Avian Influenza Neuraminidase, Tamiflu and Relenza